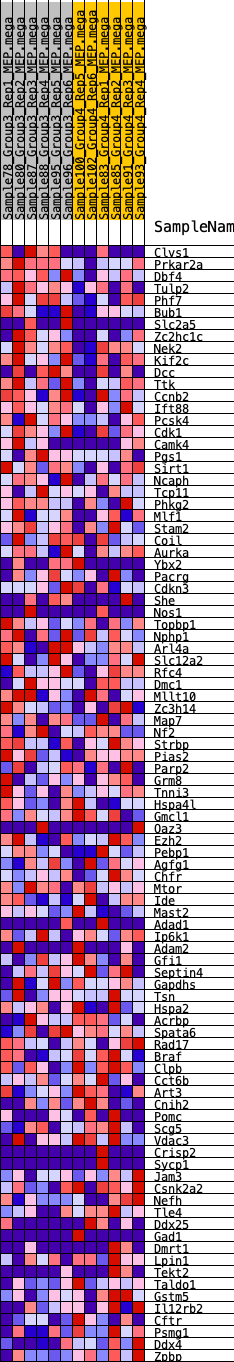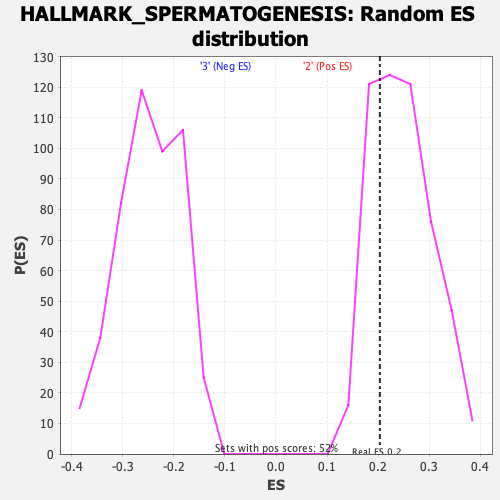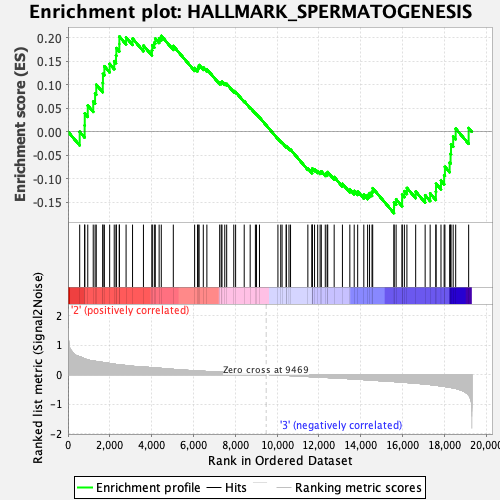
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.20357163 |
| Normalized Enrichment Score (NES) | 0.8247995 |
| Nominal p-value | 0.72868216 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Clvs1 | 559 | 0.597 | -0.0001 | Yes |
| 2 | Prkar2a | 796 | 0.530 | 0.0134 | Yes |
| 3 | Dbf4 | 802 | 0.529 | 0.0388 | Yes |
| 4 | Tulp2 | 945 | 0.497 | 0.0556 | Yes |
| 5 | Phf7 | 1206 | 0.456 | 0.0642 | Yes |
| 6 | Bub1 | 1295 | 0.448 | 0.0813 | Yes |
| 7 | Slc2a5 | 1351 | 0.442 | 0.1000 | Yes |
| 8 | Zc2hc1c | 1663 | 0.409 | 0.1037 | Yes |
| 9 | Nek2 | 1672 | 0.407 | 0.1230 | Yes |
| 10 | Kif2c | 1739 | 0.400 | 0.1390 | Yes |
| 11 | Dcc | 1992 | 0.374 | 0.1441 | Yes |
| 12 | Ttk | 2210 | 0.350 | 0.1498 | Yes |
| 13 | Ccnb2 | 2296 | 0.341 | 0.1620 | Yes |
| 14 | Ift88 | 2312 | 0.339 | 0.1777 | Yes |
| 15 | Pcsk4 | 2454 | 0.329 | 0.1863 | Yes |
| 16 | Cdk1 | 2456 | 0.329 | 0.2022 | Yes |
| 17 | Camk4 | 2777 | 0.304 | 0.2004 | Yes |
| 18 | Pgs1 | 3086 | 0.280 | 0.1979 | Yes |
| 19 | Sirt1 | 3607 | 0.249 | 0.1830 | Yes |
| 20 | Ncaph | 4013 | 0.232 | 0.1732 | Yes |
| 21 | Tcp11 | 4027 | 0.231 | 0.1837 | Yes |
| 22 | Phkg2 | 4128 | 0.225 | 0.1894 | Yes |
| 23 | Mlf1 | 4173 | 0.222 | 0.1979 | Yes |
| 24 | Stam2 | 4357 | 0.214 | 0.1988 | Yes |
| 25 | Coil | 4460 | 0.208 | 0.2036 | Yes |
| 26 | Aurka | 5035 | 0.174 | 0.1821 | No |
| 27 | Ybx2 | 6054 | 0.127 | 0.1353 | No |
| 28 | Pacrg | 6195 | 0.120 | 0.1339 | No |
| 29 | Cdkn3 | 6226 | 0.119 | 0.1381 | No |
| 30 | She | 6269 | 0.117 | 0.1416 | No |
| 31 | Nos1 | 6469 | 0.109 | 0.1366 | No |
| 32 | Topbp1 | 6640 | 0.102 | 0.1327 | No |
| 33 | Nphp1 | 7252 | 0.079 | 0.1047 | No |
| 34 | Arl4a | 7325 | 0.076 | 0.1047 | No |
| 35 | Slc12a2 | 7362 | 0.075 | 0.1065 | No |
| 36 | Rfc4 | 7495 | 0.071 | 0.1030 | No |
| 37 | Dmc1 | 7587 | 0.067 | 0.1016 | No |
| 38 | Mllt10 | 7926 | 0.053 | 0.0866 | No |
| 39 | Zc3h14 | 8014 | 0.050 | 0.0844 | No |
| 40 | Map7 | 8425 | 0.035 | 0.0648 | No |
| 41 | Nf2 | 8710 | 0.025 | 0.0512 | No |
| 42 | Strbp | 8960 | 0.018 | 0.0391 | No |
| 43 | Pias2 | 9011 | 0.016 | 0.0373 | No |
| 44 | Parp2 | 9155 | 0.011 | 0.0304 | No |
| 45 | Grm8 | 10037 | -0.014 | -0.0148 | No |
| 46 | Tnni3 | 10171 | -0.019 | -0.0208 | No |
| 47 | Hspa4l | 10235 | -0.021 | -0.0231 | No |
| 48 | Gmcl1 | 10434 | -0.028 | -0.0320 | No |
| 49 | Oaz3 | 10436 | -0.028 | -0.0307 | No |
| 50 | Ezh2 | 10565 | -0.032 | -0.0358 | No |
| 51 | Pebp1 | 10640 | -0.035 | -0.0380 | No |
| 52 | Agfg1 | 11468 | -0.065 | -0.0779 | No |
| 53 | Chfr | 11669 | -0.073 | -0.0847 | No |
| 54 | Mtor | 11671 | -0.073 | -0.0812 | No |
| 55 | Ide | 11679 | -0.073 | -0.0781 | No |
| 56 | Mast2 | 11788 | -0.077 | -0.0799 | No |
| 57 | Adad1 | 11938 | -0.083 | -0.0837 | No |
| 58 | Ip6k1 | 12054 | -0.086 | -0.0855 | No |
| 59 | Adam2 | 12111 | -0.088 | -0.0842 | No |
| 60 | Gfi1 | 12301 | -0.095 | -0.0894 | No |
| 61 | Septin4 | 12388 | -0.099 | -0.0890 | No |
| 62 | Gapdhs | 12418 | -0.100 | -0.0857 | No |
| 63 | Tsn | 12726 | -0.112 | -0.0962 | No |
| 64 | Hspa2 | 13121 | -0.126 | -0.1106 | No |
| 65 | Acrbp | 13475 | -0.140 | -0.1222 | No |
| 66 | Spata6 | 13684 | -0.148 | -0.1258 | No |
| 67 | Rad17 | 13850 | -0.156 | -0.1268 | No |
| 68 | Braf | 14143 | -0.169 | -0.1338 | No |
| 69 | Clpb | 14325 | -0.177 | -0.1346 | No |
| 70 | Cct6b | 14417 | -0.181 | -0.1305 | No |
| 71 | Art3 | 14536 | -0.187 | -0.1276 | No |
| 72 | Cnih2 | 14568 | -0.189 | -0.1200 | No |
| 73 | Pomc | 15587 | -0.236 | -0.1615 | No |
| 74 | Scg5 | 15589 | -0.236 | -0.1501 | No |
| 75 | Vdac3 | 15688 | -0.241 | -0.1435 | No |
| 76 | Crisp2 | 15975 | -0.256 | -0.1459 | No |
| 77 | Sycp1 | 15976 | -0.256 | -0.1334 | No |
| 78 | Jam3 | 16083 | -0.261 | -0.1263 | No |
| 79 | Csnk2a2 | 16204 | -0.269 | -0.1195 | No |
| 80 | Nefh | 16623 | -0.291 | -0.1271 | No |
| 81 | Tle4 | 17076 | -0.321 | -0.1350 | No |
| 82 | Ddx25 | 17315 | -0.338 | -0.1309 | No |
| 83 | Gad1 | 17588 | -0.361 | -0.1275 | No |
| 84 | Dmrt1 | 17596 | -0.362 | -0.1103 | No |
| 85 | Lpin1 | 17833 | -0.385 | -0.1039 | No |
| 86 | Tekt2 | 17984 | -0.399 | -0.0923 | No |
| 87 | Taldo1 | 18021 | -0.404 | -0.0745 | No |
| 88 | Gstm5 | 18247 | -0.425 | -0.0656 | No |
| 89 | Il12rb2 | 18292 | -0.433 | -0.0468 | No |
| 90 | Cftr | 18313 | -0.436 | -0.0267 | No |
| 91 | Psmg1 | 18416 | -0.452 | -0.0101 | No |
| 92 | Ddx4 | 18536 | -0.469 | 0.0065 | No |
| 93 | Zpbp | 19156 | -0.690 | 0.0079 | No |