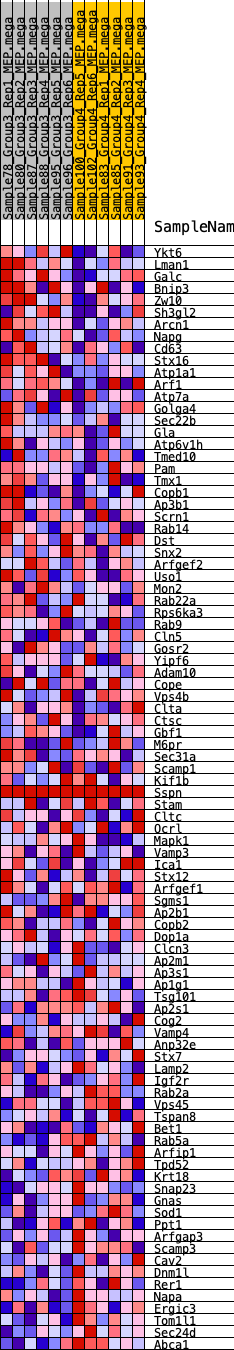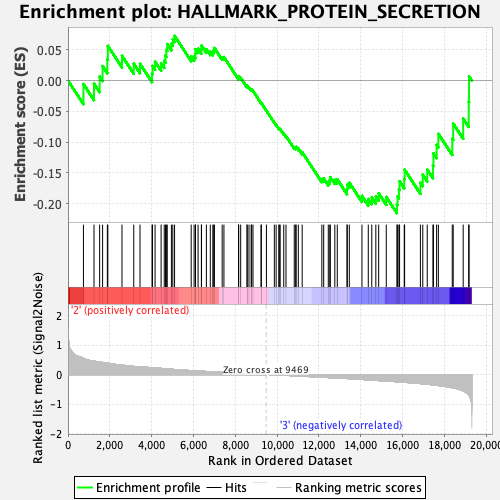
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | -0.2156882 |
| Normalized Enrichment Score (NES) | -1.0565975 |
| Nominal p-value | 0.36055776 |
| FDR q-value | 0.83770174 |
| FWER p-Value | 0.984 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ykt6 | 738 | 0.549 | -0.0057 | No |
| 2 | Lman1 | 1243 | 0.452 | -0.0050 | No |
| 3 | Galc | 1513 | 0.426 | 0.0064 | No |
| 4 | Bnip3 | 1654 | 0.410 | 0.0235 | No |
| 5 | Zw10 | 1881 | 0.387 | 0.0348 | No |
| 6 | Sh3gl2 | 1905 | 0.382 | 0.0563 | No |
| 7 | Arcn1 | 2581 | 0.320 | 0.0403 | No |
| 8 | Napg | 3144 | 0.275 | 0.0274 | No |
| 9 | Cd63 | 3442 | 0.257 | 0.0273 | No |
| 10 | Stx16 | 4017 | 0.232 | 0.0112 | No |
| 11 | Atp1a1 | 4041 | 0.230 | 0.0237 | No |
| 12 | Arf1 | 4163 | 0.223 | 0.0307 | No |
| 13 | Atp7a | 4451 | 0.208 | 0.0281 | No |
| 14 | Golga4 | 4601 | 0.198 | 0.0322 | No |
| 15 | Sec22b | 4661 | 0.196 | 0.0408 | No |
| 16 | Gla | 4704 | 0.194 | 0.0502 | No |
| 17 | Atp6v1h | 4745 | 0.191 | 0.0594 | No |
| 18 | Tmed10 | 4944 | 0.179 | 0.0598 | No |
| 19 | Pam | 5011 | 0.175 | 0.0668 | No |
| 20 | Tmx1 | 5095 | 0.170 | 0.0727 | No |
| 21 | Copb1 | 5890 | 0.132 | 0.0392 | No |
| 22 | Ap3b1 | 6029 | 0.128 | 0.0396 | No |
| 23 | Scrn1 | 6093 | 0.125 | 0.0438 | No |
| 24 | Rab14 | 6096 | 0.125 | 0.0511 | No |
| 25 | Dst | 6219 | 0.119 | 0.0519 | No |
| 26 | Snx2 | 6377 | 0.113 | 0.0505 | No |
| 27 | Arfgef2 | 6384 | 0.113 | 0.0569 | No |
| 28 | Uso1 | 6616 | 0.103 | 0.0510 | No |
| 29 | Mon2 | 6803 | 0.097 | 0.0471 | No |
| 30 | Rab22a | 6925 | 0.091 | 0.0462 | No |
| 31 | Rps6ka3 | 6968 | 0.089 | 0.0493 | No |
| 32 | Rab9 | 7000 | 0.088 | 0.0530 | No |
| 33 | Cln5 | 7371 | 0.075 | 0.0381 | No |
| 34 | Gosr2 | 7455 | 0.073 | 0.0382 | No |
| 35 | Yipf6 | 8155 | 0.044 | 0.0044 | No |
| 36 | Adam10 | 8160 | 0.044 | 0.0068 | No |
| 37 | Cope | 8256 | 0.041 | 0.0043 | No |
| 38 | Vps4b | 8560 | 0.030 | -0.0096 | No |
| 39 | Clta | 8566 | 0.030 | -0.0081 | No |
| 40 | Ctsc | 8653 | 0.027 | -0.0110 | No |
| 41 | Gbf1 | 8757 | 0.023 | -0.0149 | No |
| 42 | M6pr | 8780 | 0.022 | -0.0147 | No |
| 43 | Sec31a | 8845 | 0.020 | -0.0169 | No |
| 44 | Scamp1 | 9230 | 0.008 | -0.0364 | No |
| 45 | Kif1b | 9243 | 0.008 | -0.0365 | No |
| 46 | Sspn | 9487 | 0.000 | -0.0492 | No |
| 47 | Stam | 9865 | -0.008 | -0.0683 | No |
| 48 | Cltc | 9952 | -0.011 | -0.0721 | No |
| 49 | Ocrl | 10076 | -0.015 | -0.0776 | No |
| 50 | Mapk1 | 10123 | -0.017 | -0.0790 | No |
| 51 | Vamp3 | 10139 | -0.017 | -0.0788 | No |
| 52 | Ica1 | 10315 | -0.024 | -0.0865 | No |
| 53 | Stx12 | 10422 | -0.028 | -0.0904 | No |
| 54 | Arfgef1 | 10818 | -0.041 | -0.1085 | No |
| 55 | Sgms1 | 10886 | -0.043 | -0.1094 | No |
| 56 | Ap2b1 | 10908 | -0.044 | -0.1078 | No |
| 57 | Copb2 | 11012 | -0.048 | -0.1103 | No |
| 58 | Dop1a | 11197 | -0.055 | -0.1166 | No |
| 59 | Clcn3 | 12137 | -0.089 | -0.1602 | No |
| 60 | Ap2m1 | 12225 | -0.092 | -0.1592 | No |
| 61 | Ap3s1 | 12445 | -0.101 | -0.1646 | No |
| 62 | Ap1g1 | 12516 | -0.104 | -0.1621 | No |
| 63 | Tsg101 | 12541 | -0.105 | -0.1571 | No |
| 64 | Ap2s1 | 12757 | -0.113 | -0.1616 | No |
| 65 | Cog2 | 12875 | -0.117 | -0.1607 | No |
| 66 | Vamp4 | 13341 | -0.134 | -0.1769 | No |
| 67 | Anp32e | 13351 | -0.135 | -0.1693 | No |
| 68 | Stx7 | 13450 | -0.139 | -0.1661 | No |
| 69 | Lamp2 | 14054 | -0.166 | -0.1876 | No |
| 70 | Igf2r | 14357 | -0.179 | -0.1927 | No |
| 71 | Rab2a | 14524 | -0.186 | -0.1902 | No |
| 72 | Vps45 | 14718 | -0.197 | -0.1886 | No |
| 73 | Tspan8 | 14854 | -0.204 | -0.1835 | No |
| 74 | Bet1 | 15217 | -0.218 | -0.1893 | No |
| 75 | Rab5a | 15725 | -0.242 | -0.2013 | Yes |
| 76 | Arfip1 | 15759 | -0.243 | -0.1885 | Yes |
| 77 | Tpd52 | 15823 | -0.247 | -0.1771 | Yes |
| 78 | Krt18 | 15853 | -0.249 | -0.1638 | Yes |
| 79 | Snap23 | 16076 | -0.260 | -0.1598 | Yes |
| 80 | Gnas | 16094 | -0.262 | -0.1451 | Yes |
| 81 | Sod1 | 16849 | -0.306 | -0.1661 | Yes |
| 82 | Ppt1 | 16963 | -0.315 | -0.1532 | Yes |
| 83 | Arfgap3 | 17176 | -0.326 | -0.1448 | Yes |
| 84 | Scamp3 | 17449 | -0.350 | -0.1382 | Yes |
| 85 | Cav2 | 17472 | -0.352 | -0.1183 | Yes |
| 86 | Dnm1l | 17624 | -0.364 | -0.1045 | Yes |
| 87 | Rer1 | 17708 | -0.372 | -0.0867 | Yes |
| 88 | Napa | 18372 | -0.446 | -0.0946 | Yes |
| 89 | Ergic3 | 18417 | -0.452 | -0.0700 | Yes |
| 90 | Tom1l1 | 18894 | -0.554 | -0.0618 | Yes |
| 91 | Sec24d | 19159 | -0.693 | -0.0342 | Yes |
| 92 | Abca1 | 19175 | -0.703 | 0.0069 | Yes |