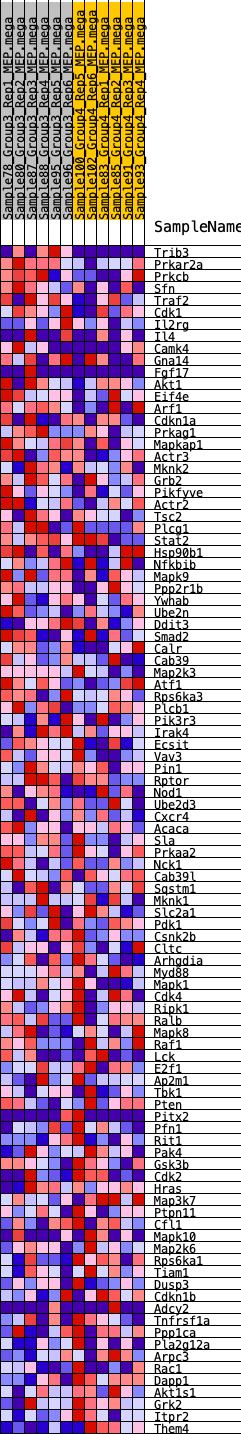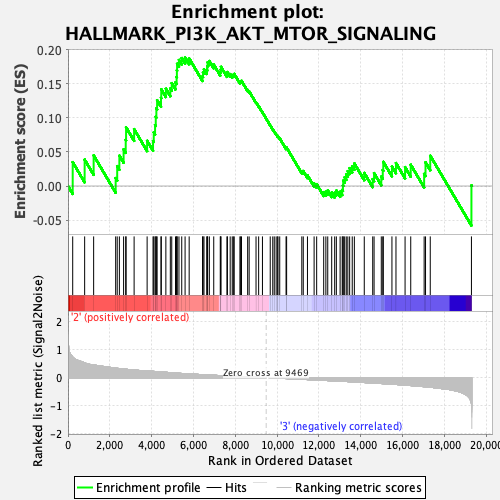
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.18849362 |
| Normalized Enrichment Score (NES) | 0.85164136 |
| Nominal p-value | 0.64615387 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Trib3 | 225 | 0.741 | 0.0350 | Yes |
| 2 | Prkar2a | 796 | 0.530 | 0.0387 | Yes |
| 3 | Prkcb | 1226 | 0.453 | 0.0449 | Yes |
| 4 | Sfn | 2279 | 0.342 | 0.0117 | Yes |
| 5 | Traf2 | 2353 | 0.336 | 0.0291 | Yes |
| 6 | Cdk1 | 2456 | 0.329 | 0.0445 | Yes |
| 7 | Il2rg | 2658 | 0.313 | 0.0538 | Yes |
| 8 | Il4 | 2760 | 0.305 | 0.0677 | Yes |
| 9 | Camk4 | 2777 | 0.304 | 0.0860 | Yes |
| 10 | Gna14 | 3161 | 0.274 | 0.0834 | Yes |
| 11 | Fgf17 | 3783 | 0.243 | 0.0663 | Yes |
| 12 | Akt1 | 4066 | 0.229 | 0.0660 | Yes |
| 13 | Eif4e | 4100 | 0.227 | 0.0786 | Yes |
| 14 | Arf1 | 4163 | 0.223 | 0.0894 | Yes |
| 15 | Cdkn1a | 4196 | 0.220 | 0.1016 | Yes |
| 16 | Prkag1 | 4225 | 0.218 | 0.1139 | Yes |
| 17 | Mapkap1 | 4261 | 0.217 | 0.1257 | Yes |
| 18 | Actr3 | 4447 | 0.208 | 0.1292 | Yes |
| 19 | Mknk2 | 4459 | 0.208 | 0.1417 | Yes |
| 20 | Grb2 | 4679 | 0.195 | 0.1426 | Yes |
| 21 | Pikfyve | 4892 | 0.182 | 0.1431 | Yes |
| 22 | Actr2 | 4960 | 0.179 | 0.1508 | Yes |
| 23 | Tsc2 | 5140 | 0.168 | 0.1521 | Yes |
| 24 | Plcg1 | 5193 | 0.165 | 0.1598 | Yes |
| 25 | Stat2 | 5206 | 0.165 | 0.1695 | Yes |
| 26 | Hsp90b1 | 5217 | 0.164 | 0.1793 | Yes |
| 27 | Nfkbib | 5305 | 0.159 | 0.1848 | Yes |
| 28 | Mapk9 | 5438 | 0.153 | 0.1876 | Yes |
| 29 | Ppp2r1b | 5598 | 0.146 | 0.1885 | Yes |
| 30 | Ywhab | 5793 | 0.137 | 0.1870 | No |
| 31 | Ube2n | 6431 | 0.111 | 0.1608 | No |
| 32 | Ddit3 | 6456 | 0.110 | 0.1665 | No |
| 33 | Smad2 | 6501 | 0.108 | 0.1710 | No |
| 34 | Calr | 6634 | 0.103 | 0.1706 | No |
| 35 | Cab39 | 6662 | 0.102 | 0.1756 | No |
| 36 | Map2k3 | 6676 | 0.101 | 0.1813 | No |
| 37 | Atf1 | 6758 | 0.098 | 0.1833 | No |
| 38 | Rps6ka3 | 6968 | 0.089 | 0.1780 | No |
| 39 | Plcb1 | 7280 | 0.078 | 0.1667 | No |
| 40 | Pik3r3 | 7300 | 0.077 | 0.1706 | No |
| 41 | Irak4 | 7313 | 0.077 | 0.1748 | No |
| 42 | Ecsit | 7600 | 0.067 | 0.1641 | No |
| 43 | Vav3 | 7622 | 0.065 | 0.1671 | No |
| 44 | Pin1 | 7754 | 0.059 | 0.1640 | No |
| 45 | Rptor | 7852 | 0.056 | 0.1625 | No |
| 46 | Nod1 | 7905 | 0.054 | 0.1632 | No |
| 47 | Ube2d3 | 7947 | 0.053 | 0.1644 | No |
| 48 | Cxcr4 | 8220 | 0.042 | 0.1529 | No |
| 49 | Acaca | 8255 | 0.041 | 0.1537 | No |
| 50 | Sla | 8295 | 0.040 | 0.1542 | No |
| 51 | Prkaa2 | 8590 | 0.029 | 0.1407 | No |
| 52 | Nck1 | 8661 | 0.027 | 0.1388 | No |
| 53 | Cab39l | 8990 | 0.017 | 0.1228 | No |
| 54 | Sqstm1 | 9115 | 0.012 | 0.1171 | No |
| 55 | Mknk1 | 9297 | 0.006 | 0.1080 | No |
| 56 | Slc2a1 | 9669 | -0.002 | 0.0888 | No |
| 57 | Pdk1 | 9790 | -0.006 | 0.0829 | No |
| 58 | Csnk2b | 9861 | -0.008 | 0.0798 | No |
| 59 | Cltc | 9952 | -0.011 | 0.0758 | No |
| 60 | Arhgdia | 10028 | -0.013 | 0.0727 | No |
| 61 | Myd88 | 10034 | -0.014 | 0.0733 | No |
| 62 | Mapk1 | 10123 | -0.017 | 0.0698 | No |
| 63 | Cdk4 | 10426 | -0.028 | 0.0558 | No |
| 64 | Ripk1 | 10453 | -0.029 | 0.0562 | No |
| 65 | Ralb | 11179 | -0.054 | 0.0219 | No |
| 66 | Mapk8 | 11248 | -0.057 | 0.0220 | No |
| 67 | Raf1 | 11449 | -0.065 | 0.0156 | No |
| 68 | Lck | 11767 | -0.076 | 0.0039 | No |
| 69 | E2f1 | 11889 | -0.081 | 0.0027 | No |
| 70 | Ap2m1 | 12225 | -0.092 | -0.0089 | No |
| 71 | Tbk1 | 12329 | -0.097 | -0.0082 | No |
| 72 | Pten | 12419 | -0.100 | -0.0065 | No |
| 73 | Pitx2 | 12608 | -0.107 | -0.0095 | No |
| 74 | Pfn1 | 12751 | -0.113 | -0.0098 | No |
| 75 | Rit1 | 12830 | -0.115 | -0.0066 | No |
| 76 | Pak4 | 13010 | -0.122 | -0.0082 | No |
| 77 | Gsk3b | 13111 | -0.126 | -0.0055 | No |
| 78 | Cdk2 | 13145 | -0.127 | 0.0007 | No |
| 79 | Hras | 13159 | -0.127 | 0.0080 | No |
| 80 | Map3k7 | 13227 | -0.130 | 0.0127 | No |
| 81 | Ptpn11 | 13304 | -0.133 | 0.0171 | No |
| 82 | Cfl1 | 13379 | -0.136 | 0.0219 | No |
| 83 | Mapk10 | 13466 | -0.140 | 0.0262 | No |
| 84 | Map2k6 | 13590 | -0.145 | 0.0289 | No |
| 85 | Rps6ka1 | 13691 | -0.148 | 0.0331 | No |
| 86 | Tiam1 | 14165 | -0.170 | 0.0192 | No |
| 87 | Dusp3 | 14569 | -0.189 | 0.0101 | No |
| 88 | Cdkn1b | 14634 | -0.192 | 0.0189 | No |
| 89 | Adcy2 | 14979 | -0.206 | 0.0140 | No |
| 90 | Tnfrsf1a | 15050 | -0.209 | 0.0235 | No |
| 91 | Ppp1ca | 15075 | -0.210 | 0.0355 | No |
| 92 | Pla2g12a | 15488 | -0.231 | 0.0286 | No |
| 93 | Arpc3 | 15681 | -0.240 | 0.0337 | No |
| 94 | Rac1 | 16116 | -0.264 | 0.0277 | No |
| 95 | Dapp1 | 16387 | -0.279 | 0.0312 | No |
| 96 | Akt1s1 | 17028 | -0.318 | 0.0179 | No |
| 97 | Grk2 | 17094 | -0.322 | 0.0348 | No |
| 98 | Itpr2 | 17320 | -0.339 | 0.0444 | No |
| 99 | Them4 | 19289 | -0.936 | 0.0009 | No |