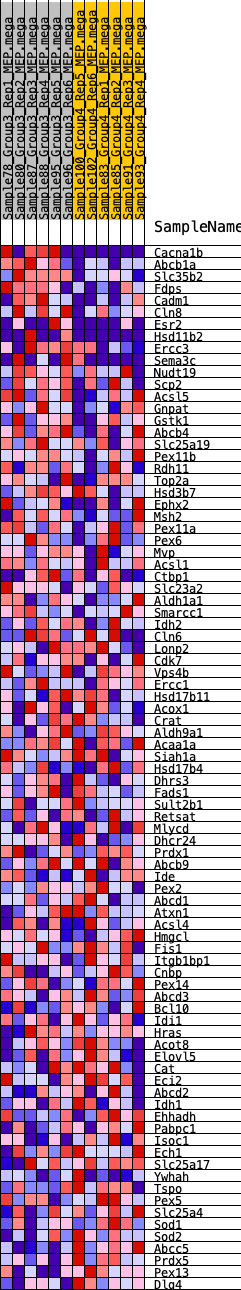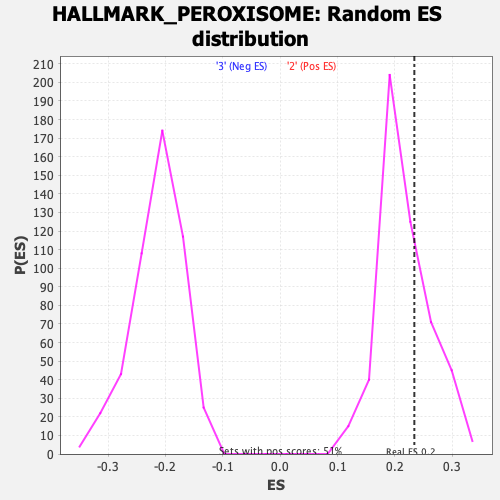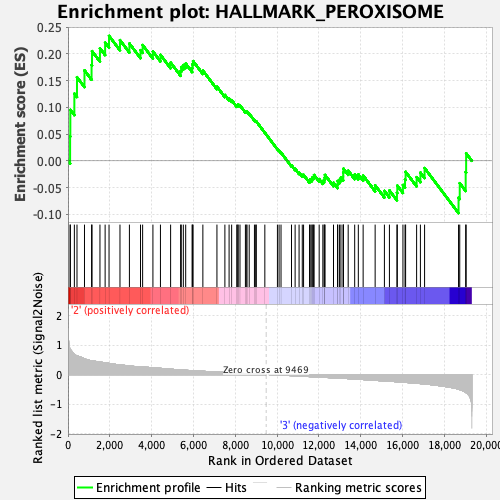
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | 0.2338815 |
| Normalized Enrichment Score (NES) | 1.0802772 |
| Nominal p-value | 0.33136094 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.97 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cacna1b | 101 | 0.869 | 0.0460 | Yes |
| 2 | Abcb1a | 112 | 0.849 | 0.0955 | Yes |
| 3 | Slc35b2 | 304 | 0.684 | 0.1258 | Yes |
| 4 | Fdps | 430 | 0.632 | 0.1566 | Yes |
| 5 | Cadm1 | 788 | 0.532 | 0.1693 | Yes |
| 6 | Cln8 | 1127 | 0.466 | 0.1792 | Yes |
| 7 | Esr2 | 1149 | 0.462 | 0.2053 | Yes |
| 8 | Hsd11b2 | 1528 | 0.424 | 0.2106 | Yes |
| 9 | Ercc3 | 1775 | 0.398 | 0.2213 | Yes |
| 10 | Sema3c | 1962 | 0.378 | 0.2339 | Yes |
| 11 | Nudt19 | 2485 | 0.326 | 0.2259 | No |
| 12 | Scp2 | 2936 | 0.291 | 0.2196 | No |
| 13 | Acsl5 | 3467 | 0.255 | 0.2071 | No |
| 14 | Gnpat | 3568 | 0.252 | 0.2168 | No |
| 15 | Gstk1 | 4060 | 0.229 | 0.2047 | No |
| 16 | Abcb4 | 4420 | 0.210 | 0.1984 | No |
| 17 | Slc25a19 | 4906 | 0.182 | 0.1838 | No |
| 18 | Pex11b | 5385 | 0.155 | 0.1681 | No |
| 19 | Rdh11 | 5415 | 0.153 | 0.1756 | No |
| 20 | Top2a | 5514 | 0.149 | 0.1793 | No |
| 21 | Hsd3b7 | 5627 | 0.144 | 0.1820 | No |
| 22 | Ephx2 | 5936 | 0.130 | 0.1737 | No |
| 23 | Msh2 | 5948 | 0.130 | 0.1807 | No |
| 24 | Pex11a | 5981 | 0.129 | 0.1867 | No |
| 25 | Pex6 | 6448 | 0.110 | 0.1689 | No |
| 26 | Mvp | 7121 | 0.083 | 0.1388 | No |
| 27 | Acsl1 | 7501 | 0.071 | 0.1233 | No |
| 28 | Ctbp1 | 7702 | 0.061 | 0.1165 | No |
| 29 | Slc23a2 | 7824 | 0.057 | 0.1136 | No |
| 30 | Aldh1a1 | 8068 | 0.048 | 0.1038 | No |
| 31 | Smarcc1 | 8116 | 0.046 | 0.1040 | No |
| 32 | Idh2 | 8136 | 0.045 | 0.1057 | No |
| 33 | Cln6 | 8223 | 0.042 | 0.1037 | No |
| 34 | Lonp2 | 8482 | 0.033 | 0.0922 | No |
| 35 | Cdk7 | 8520 | 0.031 | 0.0921 | No |
| 36 | Vps4b | 8560 | 0.030 | 0.0919 | No |
| 37 | Ercc1 | 8664 | 0.027 | 0.0881 | No |
| 38 | Hsd17b11 | 8914 | 0.019 | 0.0763 | No |
| 39 | Acox1 | 8974 | 0.017 | 0.0743 | No |
| 40 | Crat | 9003 | 0.016 | 0.0737 | No |
| 41 | Aldh9a1 | 9411 | 0.002 | 0.0527 | No |
| 42 | Acaa1a | 10013 | -0.013 | 0.0222 | No |
| 43 | Siah1a | 10092 | -0.016 | 0.0191 | No |
| 44 | Hsd17b4 | 10185 | -0.019 | 0.0154 | No |
| 45 | Dhrs3 | 10686 | -0.036 | -0.0085 | No |
| 46 | Fads1 | 10859 | -0.042 | -0.0149 | No |
| 47 | Sult2b1 | 11048 | -0.049 | -0.0218 | No |
| 48 | Retsat | 11201 | -0.055 | -0.0265 | No |
| 49 | Mlycd | 11263 | -0.057 | -0.0263 | No |
| 50 | Dhcr24 | 11548 | -0.068 | -0.0370 | No |
| 51 | Prdx1 | 11580 | -0.069 | -0.0346 | No |
| 52 | Abcb9 | 11659 | -0.072 | -0.0344 | No |
| 53 | Ide | 11679 | -0.073 | -0.0310 | No |
| 54 | Pex2 | 11742 | -0.075 | -0.0298 | No |
| 55 | Abcd1 | 11765 | -0.076 | -0.0265 | No |
| 56 | Atxn1 | 12009 | -0.084 | -0.0342 | No |
| 57 | Acsl4 | 12185 | -0.091 | -0.0379 | No |
| 58 | Hmgcl | 12244 | -0.093 | -0.0355 | No |
| 59 | Fis1 | 12273 | -0.094 | -0.0314 | No |
| 60 | Itgb1bp1 | 12285 | -0.095 | -0.0264 | No |
| 61 | Cnbp | 12694 | -0.111 | -0.0411 | No |
| 62 | Pex14 | 12889 | -0.118 | -0.0442 | No |
| 63 | Abcd3 | 12897 | -0.118 | -0.0376 | No |
| 64 | Bcl10 | 12985 | -0.121 | -0.0350 | No |
| 65 | Idi1 | 13050 | -0.123 | -0.0311 | No |
| 66 | Hras | 13159 | -0.127 | -0.0292 | No |
| 67 | Acot8 | 13160 | -0.127 | -0.0217 | No |
| 68 | Elovl5 | 13169 | -0.127 | -0.0147 | No |
| 69 | Cat | 13396 | -0.137 | -0.0184 | No |
| 70 | Eci2 | 13707 | -0.149 | -0.0257 | No |
| 71 | Abcd2 | 13881 | -0.157 | -0.0254 | No |
| 72 | Idh1 | 14111 | -0.168 | -0.0275 | No |
| 73 | Ehhadh | 14685 | -0.194 | -0.0458 | No |
| 74 | Pabpc1 | 15128 | -0.213 | -0.0563 | No |
| 75 | Isoc1 | 15368 | -0.225 | -0.0554 | No |
| 76 | Ech1 | 15728 | -0.242 | -0.0599 | No |
| 77 | Slc25a17 | 15744 | -0.243 | -0.0463 | No |
| 78 | Ywhah | 16014 | -0.257 | -0.0452 | No |
| 79 | Tspo | 16114 | -0.263 | -0.0348 | No |
| 80 | Pex5 | 16142 | -0.265 | -0.0206 | No |
| 81 | Slc25a4 | 16668 | -0.294 | -0.0306 | No |
| 82 | Sod1 | 16849 | -0.306 | -0.0220 | No |
| 83 | Sod2 | 17049 | -0.319 | -0.0135 | No |
| 84 | Abcc5 | 18673 | -0.493 | -0.0689 | No |
| 85 | Prdx5 | 18730 | -0.508 | -0.0419 | No |
| 86 | Pex13 | 19014 | -0.604 | -0.0210 | No |
| 87 | Dlg4 | 19036 | -0.614 | 0.0141 | No |