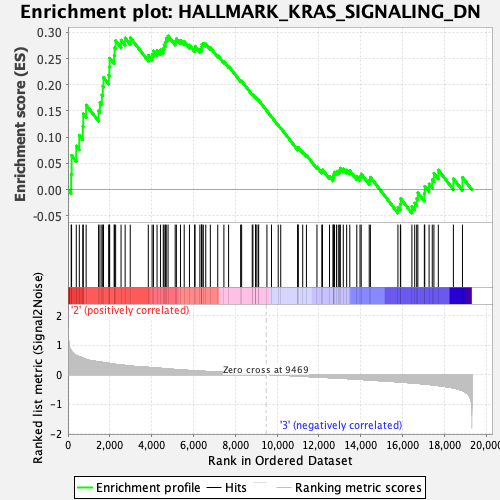
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.29325363 |
| Normalized Enrichment Score (NES) | 1.2249881 |
| Nominal p-value | 0.06944445 |
| FDR q-value | 0.7317742 |
| FWER p-Value | 0.871 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atp4a | 153 | 0.813 | 0.0294 | Yes |
| 2 | Gtf3c5 | 171 | 0.801 | 0.0654 | Yes |
| 3 | Stag3 | 396 | 0.644 | 0.0833 | Yes |
| 4 | Camk1d | 541 | 0.604 | 0.1036 | Yes |
| 5 | Pde6b | 708 | 0.558 | 0.1206 | Yes |
| 6 | Abcg4 | 729 | 0.552 | 0.1450 | Yes |
| 7 | Tfcp2l1 | 867 | 0.512 | 0.1614 | Yes |
| 8 | Lgals7 | 1462 | 0.431 | 0.1503 | Yes |
| 9 | Hsd11b2 | 1528 | 0.424 | 0.1664 | Yes |
| 10 | Cpa2 | 1620 | 0.412 | 0.1806 | Yes |
| 11 | Zc2hc1c | 1663 | 0.409 | 0.1972 | Yes |
| 12 | Slc12a3 | 1698 | 0.405 | 0.2140 | Yes |
| 13 | Brdt | 1946 | 0.379 | 0.2186 | Yes |
| 14 | Prodh | 1980 | 0.376 | 0.2342 | Yes |
| 15 | Dcc | 1992 | 0.374 | 0.2508 | Yes |
| 16 | Gpr19 | 2206 | 0.351 | 0.2559 | Yes |
| 17 | Cyp39a1 | 2229 | 0.348 | 0.2707 | Yes |
| 18 | Efhd1 | 2278 | 0.342 | 0.2840 | Yes |
| 19 | Macroh2a2 | 2537 | 0.323 | 0.2854 | Yes |
| 20 | Kcnn1 | 2732 | 0.307 | 0.2894 | Yes |
| 21 | Skil | 2976 | 0.287 | 0.2900 | Yes |
| 22 | Bard1 | 3839 | 0.242 | 0.2562 | Yes |
| 23 | Cd80 | 4020 | 0.232 | 0.2575 | Yes |
| 24 | Clstn3 | 4085 | 0.228 | 0.2647 | Yes |
| 25 | Nr6a1 | 4258 | 0.217 | 0.2657 | Yes |
| 26 | Msh5 | 4421 | 0.210 | 0.2669 | Yes |
| 27 | Tgfb2 | 4554 | 0.201 | 0.2693 | Yes |
| 28 | Ryr1 | 4596 | 0.199 | 0.2763 | Yes |
| 29 | Tent5c | 4666 | 0.196 | 0.2817 | Yes |
| 30 | Egf | 4695 | 0.194 | 0.2892 | Yes |
| 31 | Entpd7 | 4784 | 0.188 | 0.2933 | Yes |
| 32 | Epha5 | 5124 | 0.169 | 0.2834 | No |
| 33 | Itgb1bp2 | 5182 | 0.166 | 0.2880 | No |
| 34 | Idua | 5378 | 0.156 | 0.2850 | No |
| 35 | Sgk1 | 5556 | 0.148 | 0.2826 | No |
| 36 | Selenop | 5807 | 0.136 | 0.2758 | No |
| 37 | Ybx2 | 6054 | 0.127 | 0.2689 | No |
| 38 | Arpp21 | 6073 | 0.126 | 0.2737 | No |
| 39 | Kcnd1 | 6305 | 0.116 | 0.2670 | No |
| 40 | Tgm1 | 6380 | 0.113 | 0.2684 | No |
| 41 | Lfng | 6392 | 0.112 | 0.2730 | No |
| 42 | Asb7 | 6404 | 0.112 | 0.2776 | No |
| 43 | Nos1 | 6469 | 0.109 | 0.2793 | No |
| 44 | Mefv | 6586 | 0.104 | 0.2780 | No |
| 45 | Slc6a3 | 6805 | 0.097 | 0.2711 | No |
| 46 | Ngb | 7159 | 0.081 | 0.2565 | No |
| 47 | Capn9 | 7454 | 0.073 | 0.2445 | No |
| 48 | Sptbn2 | 7678 | 0.063 | 0.2358 | No |
| 49 | Plag1 | 8254 | 0.041 | 0.2077 | No |
| 50 | Nr4a2 | 8302 | 0.039 | 0.2071 | No |
| 51 | Celsr2 | 8809 | 0.021 | 0.1817 | No |
| 52 | Dtnb | 8841 | 0.020 | 0.1811 | No |
| 53 | Zfp112 | 8968 | 0.018 | 0.1753 | No |
| 54 | Sidt1 | 8984 | 0.017 | 0.1753 | No |
| 55 | Slc16a7 | 8985 | 0.017 | 0.1761 | No |
| 56 | Kmt2d | 9088 | 0.013 | 0.1714 | No |
| 57 | Thnsl2 | 9116 | 0.012 | 0.1705 | No |
| 58 | Synpo | 9511 | 0.000 | 0.1500 | No |
| 59 | Myo15a | 9730 | -0.004 | 0.1388 | No |
| 60 | Cpeb3 | 10051 | -0.014 | 0.1228 | No |
| 61 | Tnni3 | 10171 | -0.019 | 0.1175 | No |
| 62 | Tg | 10984 | -0.047 | 0.0774 | No |
| 63 | Htr1d | 10988 | -0.047 | 0.0794 | No |
| 64 | Btg2 | 11007 | -0.048 | 0.0806 | No |
| 65 | Zbtb16 | 11222 | -0.056 | 0.0721 | No |
| 66 | Cdkal1 | 11399 | -0.062 | 0.0658 | No |
| 67 | Bmpr1b | 11903 | -0.081 | 0.0433 | No |
| 68 | Sphk2 | 12136 | -0.089 | 0.0353 | No |
| 69 | Prkn | 12168 | -0.090 | 0.0378 | No |
| 70 | Mast3 | 12502 | -0.103 | 0.0252 | No |
| 71 | Slc25a23 | 12673 | -0.110 | 0.0214 | No |
| 72 | Vps50 | 12683 | -0.110 | 0.0260 | No |
| 73 | Htr1b | 12712 | -0.111 | 0.0297 | No |
| 74 | Ypel1 | 12742 | -0.112 | 0.0333 | No |
| 75 | Tenm2 | 12835 | -0.116 | 0.0339 | No |
| 76 | Fggy | 12922 | -0.119 | 0.0349 | No |
| 77 | Copz2 | 12991 | -0.121 | 0.0369 | No |
| 78 | Pdk2 | 13020 | -0.122 | 0.0411 | No |
| 79 | Nrip2 | 13162 | -0.127 | 0.0396 | No |
| 80 | Mx2 | 13323 | -0.133 | 0.0374 | No |
| 81 | Fgfr3 | 13473 | -0.140 | 0.0361 | No |
| 82 | Tex15 | 13808 | -0.154 | 0.0257 | No |
| 83 | Edar | 13962 | -0.161 | 0.0252 | No |
| 84 | Thrb | 14019 | -0.164 | 0.0298 | No |
| 85 | Ccdc106 | 14402 | -0.181 | 0.0182 | No |
| 86 | Ryr2 | 14458 | -0.183 | 0.0238 | No |
| 87 | P2rx6 | 15775 | -0.244 | -0.0335 | No |
| 88 | Grid2 | 15892 | -0.252 | -0.0280 | No |
| 89 | Snn | 15906 | -0.252 | -0.0171 | No |
| 90 | Mthfr | 16443 | -0.282 | -0.0320 | No |
| 91 | Col2a1 | 16577 | -0.289 | -0.0256 | No |
| 92 | Ptprj | 16672 | -0.294 | -0.0170 | No |
| 93 | Serpinb2 | 16727 | -0.298 | -0.0061 | No |
| 94 | Mfsd6 | 17037 | -0.319 | -0.0075 | No |
| 95 | Slc29a3 | 17057 | -0.320 | 0.0062 | No |
| 96 | Coq8a | 17264 | -0.333 | 0.0108 | No |
| 97 | Gamt | 17416 | -0.347 | 0.0189 | No |
| 98 | Klk8 | 17495 | -0.355 | 0.0312 | No |
| 99 | Gp1ba | 17706 | -0.372 | 0.0374 | No |
| 100 | Tcf7l1 | 18426 | -0.454 | 0.0208 | No |
| 101 | Rsad2 | 18860 | -0.544 | 0.0233 | No |

