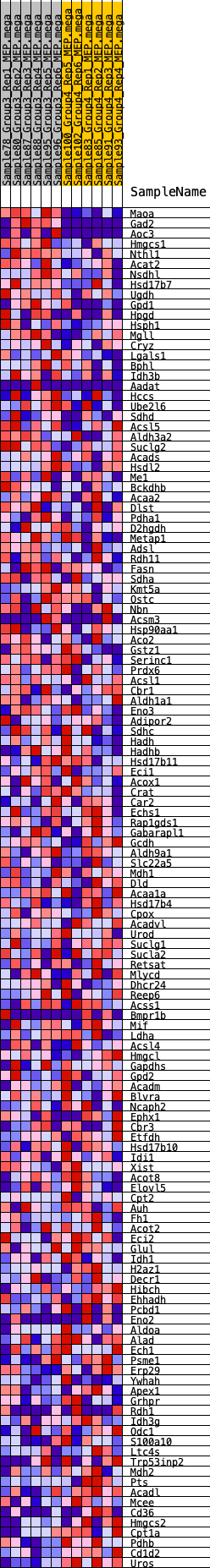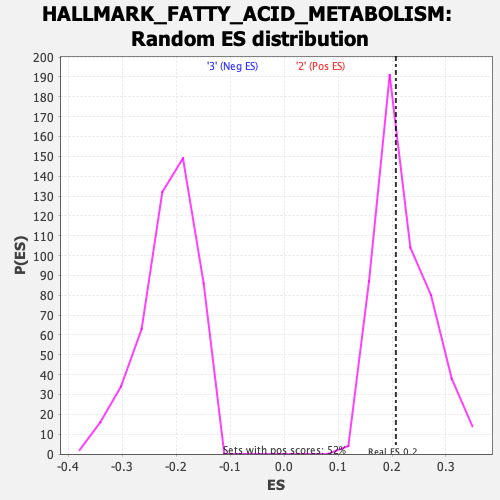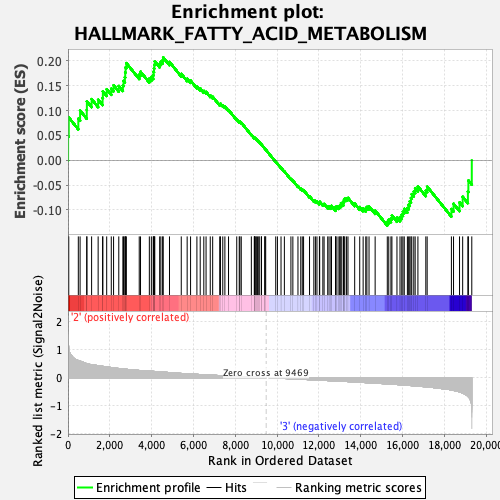
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | 0.20713785 |
| Normalized Enrichment Score (NES) | 0.9371755 |
| Nominal p-value | 0.53474903 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Maoa | 5 | 1.407 | 0.0488 | Yes |
| 2 | Gad2 | 36 | 1.115 | 0.0861 | Yes |
| 3 | Aoc3 | 493 | 0.617 | 0.0839 | Yes |
| 4 | Hmgcs1 | 578 | 0.595 | 0.1002 | Yes |
| 5 | Nthl1 | 897 | 0.507 | 0.1013 | Yes |
| 6 | Acat2 | 903 | 0.506 | 0.1187 | Yes |
| 7 | Nsdhl | 1129 | 0.465 | 0.1232 | Yes |
| 8 | Hsd17b7 | 1441 | 0.434 | 0.1221 | Yes |
| 9 | Ugdh | 1653 | 0.410 | 0.1254 | Yes |
| 10 | Gpd1 | 1667 | 0.408 | 0.1389 | Yes |
| 11 | Hpgd | 1850 | 0.390 | 0.1431 | Yes |
| 12 | Hsph1 | 2069 | 0.364 | 0.1444 | Yes |
| 13 | Mgll | 2179 | 0.354 | 0.1510 | Yes |
| 14 | Cryz | 2428 | 0.331 | 0.1496 | Yes |
| 15 | Lgals1 | 2624 | 0.315 | 0.1504 | Yes |
| 16 | Bphl | 2655 | 0.313 | 0.1598 | Yes |
| 17 | Idh3b | 2720 | 0.309 | 0.1672 | Yes |
| 18 | Aadat | 2736 | 0.307 | 0.1772 | Yes |
| 19 | Hccs | 2752 | 0.305 | 0.1870 | Yes |
| 20 | Ube2l6 | 2790 | 0.303 | 0.1957 | Yes |
| 21 | Sdhd | 3406 | 0.260 | 0.1726 | Yes |
| 22 | Acsl5 | 3467 | 0.255 | 0.1784 | Yes |
| 23 | Aldh3a2 | 3889 | 0.239 | 0.1648 | Yes |
| 24 | Suclg2 | 3985 | 0.234 | 0.1680 | Yes |
| 25 | Acads | 4074 | 0.228 | 0.1714 | Yes |
| 26 | Hsdl2 | 4089 | 0.227 | 0.1786 | Yes |
| 27 | Me1 | 4112 | 0.226 | 0.1853 | Yes |
| 28 | Bckdhb | 4129 | 0.225 | 0.1923 | Yes |
| 29 | Acaa2 | 4149 | 0.224 | 0.1991 | Yes |
| 30 | Dlst | 4383 | 0.212 | 0.1944 | Yes |
| 31 | Pdha1 | 4440 | 0.209 | 0.1987 | Yes |
| 32 | D2hgdh | 4524 | 0.204 | 0.2015 | Yes |
| 33 | Metap1 | 4552 | 0.202 | 0.2071 | Yes |
| 34 | Adsl | 4852 | 0.184 | 0.1980 | No |
| 35 | Rdh11 | 5415 | 0.153 | 0.1740 | No |
| 36 | Fasn | 5697 | 0.141 | 0.1643 | No |
| 37 | Sdha | 5860 | 0.134 | 0.1605 | No |
| 38 | Kmt5a | 6163 | 0.122 | 0.1490 | No |
| 39 | Ostc | 6321 | 0.115 | 0.1448 | No |
| 40 | Nbn | 6493 | 0.109 | 0.1397 | No |
| 41 | Acsm3 | 6601 | 0.104 | 0.1377 | No |
| 42 | Hsp90aa1 | 6801 | 0.097 | 0.1307 | No |
| 43 | Aco2 | 6917 | 0.091 | 0.1279 | No |
| 44 | Gstz1 | 7255 | 0.079 | 0.1131 | No |
| 45 | Serinc1 | 7289 | 0.077 | 0.1140 | No |
| 46 | Prdx6 | 7400 | 0.074 | 0.1109 | No |
| 47 | Acsl1 | 7501 | 0.071 | 0.1081 | No |
| 48 | Cbr1 | 7675 | 0.063 | 0.1013 | No |
| 49 | Aldh1a1 | 8068 | 0.048 | 0.0825 | No |
| 50 | Eno3 | 8187 | 0.044 | 0.0779 | No |
| 51 | Adipor2 | 8218 | 0.042 | 0.0778 | No |
| 52 | Sdhc | 8284 | 0.040 | 0.0758 | No |
| 53 | Hadh | 8766 | 0.023 | 0.0515 | No |
| 54 | Hadhb | 8909 | 0.020 | 0.0448 | No |
| 55 | Hsd17b11 | 8914 | 0.019 | 0.0453 | No |
| 56 | Eci1 | 8922 | 0.019 | 0.0456 | No |
| 57 | Acox1 | 8974 | 0.017 | 0.0435 | No |
| 58 | Crat | 9003 | 0.016 | 0.0426 | No |
| 59 | Car2 | 9057 | 0.014 | 0.0403 | No |
| 60 | Echs1 | 9124 | 0.012 | 0.0373 | No |
| 61 | Rap1gds1 | 9141 | 0.011 | 0.0369 | No |
| 62 | Gabarapl1 | 9241 | 0.008 | 0.0320 | No |
| 63 | Gcdh | 9252 | 0.007 | 0.0317 | No |
| 64 | Aldh9a1 | 9411 | 0.002 | 0.0235 | No |
| 65 | Slc22a5 | 9413 | 0.002 | 0.0235 | No |
| 66 | Mdh1 | 9452 | 0.001 | 0.0216 | No |
| 67 | Dld | 9932 | -0.010 | -0.0030 | No |
| 68 | Acaa1a | 10013 | -0.013 | -0.0067 | No |
| 69 | Hsd17b4 | 10185 | -0.019 | -0.0150 | No |
| 70 | Cpox | 10345 | -0.024 | -0.0224 | No |
| 71 | Acadvl | 10658 | -0.035 | -0.0375 | No |
| 72 | Urod | 10751 | -0.039 | -0.0409 | No |
| 73 | Suclg1 | 10995 | -0.047 | -0.0519 | No |
| 74 | Sucla2 | 11129 | -0.052 | -0.0570 | No |
| 75 | Retsat | 11201 | -0.055 | -0.0588 | No |
| 76 | Mlycd | 11263 | -0.057 | -0.0600 | No |
| 77 | Dhcr24 | 11548 | -0.068 | -0.0724 | No |
| 78 | Reep6 | 11754 | -0.076 | -0.0805 | No |
| 79 | Acss1 | 11829 | -0.078 | -0.0816 | No |
| 80 | Bmpr1b | 11903 | -0.081 | -0.0826 | No |
| 81 | Mif | 12020 | -0.085 | -0.0857 | No |
| 82 | Ldha | 12034 | -0.085 | -0.0834 | No |
| 83 | Acsl4 | 12185 | -0.091 | -0.0881 | No |
| 84 | Hmgcl | 12244 | -0.093 | -0.0879 | No |
| 85 | Gapdhs | 12418 | -0.100 | -0.0934 | No |
| 86 | Gpd2 | 12481 | -0.102 | -0.0930 | No |
| 87 | Acadm | 12569 | -0.106 | -0.0939 | No |
| 88 | Blvra | 12598 | -0.107 | -0.0916 | No |
| 89 | Ncaph2 | 12793 | -0.115 | -0.0977 | No |
| 90 | Ephx1 | 12795 | -0.115 | -0.0938 | No |
| 91 | Cbr3 | 12857 | -0.117 | -0.0929 | No |
| 92 | Etfdh | 12954 | -0.120 | -0.0937 | No |
| 93 | Hsd17b10 | 12993 | -0.121 | -0.0915 | No |
| 94 | Idi1 | 13050 | -0.123 | -0.0901 | No |
| 95 | Xist | 13056 | -0.124 | -0.0860 | No |
| 96 | Acot8 | 13160 | -0.127 | -0.0870 | No |
| 97 | Elovl5 | 13169 | -0.127 | -0.0830 | No |
| 98 | Cpt2 | 13197 | -0.128 | -0.0799 | No |
| 99 | Auh | 13234 | -0.130 | -0.0772 | No |
| 100 | Fh1 | 13320 | -0.133 | -0.0770 | No |
| 101 | Acot2 | 13386 | -0.136 | -0.0756 | No |
| 102 | Eci2 | 13707 | -0.149 | -0.0871 | No |
| 103 | Glul | 13948 | -0.160 | -0.0941 | No |
| 104 | Idh1 | 14111 | -0.168 | -0.0967 | No |
| 105 | H2az1 | 14237 | -0.173 | -0.0971 | No |
| 106 | Decr1 | 14290 | -0.175 | -0.0937 | No |
| 107 | Hibch | 14393 | -0.180 | -0.0928 | No |
| 108 | Ehhadh | 14685 | -0.194 | -0.1012 | No |
| 109 | Pcbd1 | 15266 | -0.221 | -0.1237 | No |
| 110 | Eno2 | 15336 | -0.223 | -0.1195 | No |
| 111 | Aldoa | 15440 | -0.229 | -0.1169 | No |
| 112 | Alad | 15487 | -0.231 | -0.1113 | No |
| 113 | Ech1 | 15728 | -0.242 | -0.1154 | No |
| 114 | Psme1 | 15874 | -0.251 | -0.1142 | No |
| 115 | Erp29 | 15958 | -0.255 | -0.1096 | No |
| 116 | Ywhah | 16014 | -0.257 | -0.1035 | No |
| 117 | Apex1 | 16077 | -0.261 | -0.0976 | No |
| 118 | Grhpr | 16227 | -0.270 | -0.0960 | No |
| 119 | Rdh1 | 16287 | -0.274 | -0.0895 | No |
| 120 | Idh3g | 16340 | -0.275 | -0.0826 | No |
| 121 | Odc1 | 16398 | -0.279 | -0.0759 | No |
| 122 | S100a10 | 16438 | -0.282 | -0.0681 | No |
| 123 | Ltc4s | 16533 | -0.287 | -0.0630 | No |
| 124 | Trp53inp2 | 16598 | -0.290 | -0.0562 | No |
| 125 | Mdh2 | 16730 | -0.299 | -0.0526 | No |
| 126 | Pts | 17108 | -0.323 | -0.0610 | No |
| 127 | Acadl | 17172 | -0.326 | -0.0530 | No |
| 128 | Mcee | 18331 | -0.439 | -0.0980 | No |
| 129 | Cd36 | 18433 | -0.455 | -0.0874 | No |
| 130 | Hmgcs2 | 18720 | -0.506 | -0.0847 | No |
| 131 | Cpt1a | 18870 | -0.547 | -0.0734 | No |
| 132 | Pdhb | 19120 | -0.664 | -0.0632 | No |
| 133 | Cd1d2 | 19143 | -0.680 | -0.0406 | No |
| 134 | Uros | 19306 | -1.408 | 0.0001 | No |