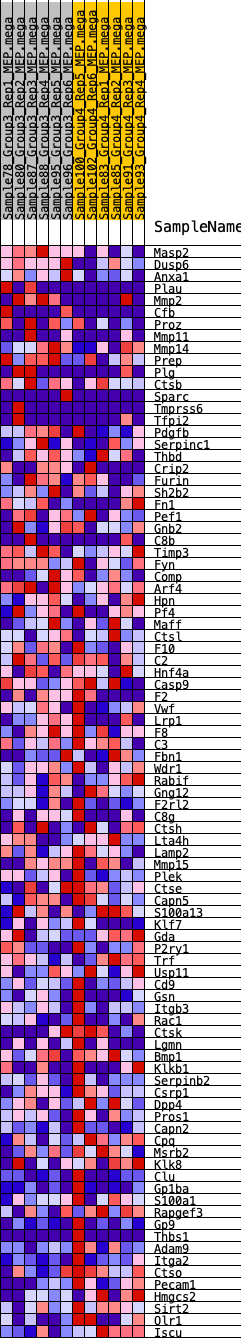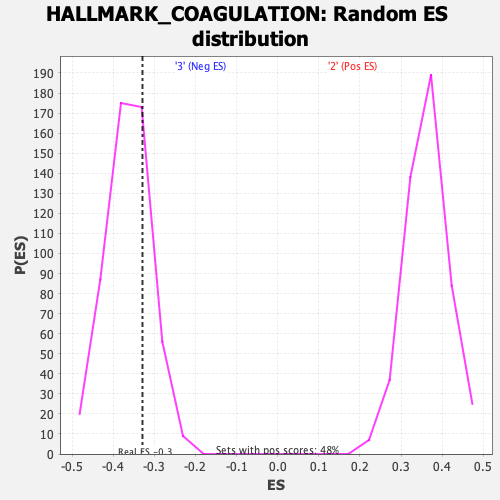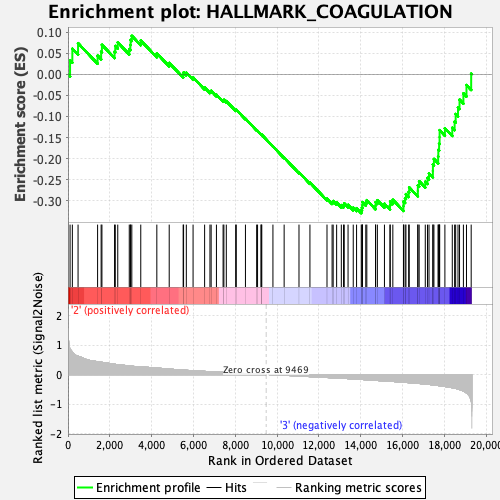
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | -0.32895 |
| Normalized Enrichment Score (NES) | -0.90541464 |
| Nominal p-value | 0.7480769 |
| FDR q-value | 0.9983823 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Masp2 | 99 | 0.872 | 0.0330 | No |
| 2 | Dusp6 | 208 | 0.755 | 0.0605 | No |
| 3 | Anxa1 | 481 | 0.621 | 0.0735 | No |
| 4 | Plau | 1415 | 0.437 | 0.0440 | No |
| 5 | Mmp2 | 1584 | 0.418 | 0.0536 | No |
| 6 | Cfb | 1622 | 0.412 | 0.0697 | No |
| 7 | Proz | 2226 | 0.348 | 0.0536 | No |
| 8 | Mmp11 | 2260 | 0.345 | 0.0669 | No |
| 9 | Mmp14 | 2379 | 0.335 | 0.0754 | No |
| 10 | Prep | 2929 | 0.291 | 0.0596 | No |
| 11 | Plg | 2981 | 0.287 | 0.0695 | No |
| 12 | Ctsb | 2995 | 0.287 | 0.0814 | No |
| 13 | Sparc | 3051 | 0.282 | 0.0909 | No |
| 14 | Tmprss6 | 3479 | 0.255 | 0.0799 | No |
| 15 | Tfpi2 | 4247 | 0.217 | 0.0494 | No |
| 16 | Pdgfb | 4839 | 0.185 | 0.0268 | No |
| 17 | Serpinc1 | 5506 | 0.149 | -0.0013 | No |
| 18 | Thbd | 5526 | 0.149 | 0.0042 | No |
| 19 | Crip2 | 5657 | 0.143 | 0.0037 | No |
| 20 | Furin | 5980 | 0.129 | -0.0074 | No |
| 21 | Sh2b2 | 6532 | 0.107 | -0.0314 | No |
| 22 | Fn1 | 6787 | 0.097 | -0.0404 | No |
| 23 | Pef1 | 6844 | 0.095 | -0.0391 | No |
| 24 | Gnb2 | 7097 | 0.084 | -0.0486 | No |
| 25 | C8b | 7415 | 0.073 | -0.0618 | No |
| 26 | Timp3 | 7453 | 0.073 | -0.0606 | No |
| 27 | Fyn | 7569 | 0.068 | -0.0636 | No |
| 28 | Comp | 8017 | 0.050 | -0.0847 | No |
| 29 | Arf4 | 8045 | 0.049 | -0.0840 | No |
| 30 | Hpn | 8484 | 0.033 | -0.1053 | No |
| 31 | Pf4 | 9020 | 0.015 | -0.1325 | No |
| 32 | Maff | 9063 | 0.014 | -0.1341 | No |
| 33 | Ctsl | 9233 | 0.008 | -0.1425 | No |
| 34 | F10 | 9236 | 0.008 | -0.1423 | No |
| 35 | C2 | 9265 | 0.007 | -0.1434 | No |
| 36 | Hnf4a | 9798 | -0.006 | -0.1709 | No |
| 37 | Casp9 | 10334 | -0.024 | -0.1976 | No |
| 38 | F2 | 11044 | -0.049 | -0.2324 | No |
| 39 | Vwf | 11567 | -0.069 | -0.2565 | No |
| 40 | Lrp1 | 12383 | -0.099 | -0.2946 | No |
| 41 | F8 | 12627 | -0.108 | -0.3025 | No |
| 42 | C3 | 12678 | -0.110 | -0.3003 | No |
| 43 | Fbn1 | 12844 | -0.116 | -0.3038 | No |
| 44 | Wdr1 | 13068 | -0.124 | -0.3100 | No |
| 45 | Rabif | 13174 | -0.128 | -0.3099 | No |
| 46 | Gng12 | 13207 | -0.129 | -0.3059 | No |
| 47 | F2rl2 | 13390 | -0.136 | -0.3094 | No |
| 48 | C8g | 13638 | -0.147 | -0.3158 | No |
| 49 | Ctsh | 13797 | -0.153 | -0.3173 | No |
| 50 | Lta4h | 14021 | -0.164 | -0.3218 | Yes |
| 51 | Lamp2 | 14054 | -0.166 | -0.3162 | Yes |
| 52 | Mmp15 | 14077 | -0.167 | -0.3101 | Yes |
| 53 | Plek | 14080 | -0.167 | -0.3028 | Yes |
| 54 | Ctse | 14238 | -0.173 | -0.3034 | Yes |
| 55 | Capn5 | 14287 | -0.175 | -0.2983 | Yes |
| 56 | S100a13 | 14704 | -0.195 | -0.3114 | Yes |
| 57 | Klf7 | 14707 | -0.196 | -0.3029 | Yes |
| 58 | Gda | 14791 | -0.200 | -0.2984 | Yes |
| 59 | P2ry1 | 15130 | -0.213 | -0.3067 | Yes |
| 60 | Trf | 15395 | -0.226 | -0.3105 | Yes |
| 61 | Usp11 | 15405 | -0.227 | -0.3011 | Yes |
| 62 | Cd9 | 15531 | -0.233 | -0.2974 | Yes |
| 63 | Gsn | 16038 | -0.258 | -0.3124 | Yes |
| 64 | Itgb3 | 16042 | -0.258 | -0.3013 | Yes |
| 65 | Rac1 | 16116 | -0.264 | -0.2935 | Yes |
| 66 | Ctsk | 16157 | -0.266 | -0.2840 | Yes |
| 67 | Lgmn | 16289 | -0.274 | -0.2788 | Yes |
| 68 | Bmp1 | 16314 | -0.274 | -0.2681 | Yes |
| 69 | Klkb1 | 16726 | -0.298 | -0.2764 | Yes |
| 70 | Serpinb2 | 16727 | -0.298 | -0.2633 | Yes |
| 71 | Csrp1 | 16789 | -0.302 | -0.2533 | Yes |
| 72 | Dpp4 | 17082 | -0.321 | -0.2544 | Yes |
| 73 | Pros1 | 17188 | -0.327 | -0.2456 | Yes |
| 74 | Capn2 | 17260 | -0.333 | -0.2347 | Yes |
| 75 | Cpq | 17446 | -0.349 | -0.2290 | Yes |
| 76 | Msrb2 | 17448 | -0.350 | -0.2138 | Yes |
| 77 | Klk8 | 17495 | -0.355 | -0.2006 | Yes |
| 78 | Clu | 17698 | -0.371 | -0.1949 | Yes |
| 79 | Gp1ba | 17706 | -0.372 | -0.1790 | Yes |
| 80 | S100a1 | 17744 | -0.376 | -0.1644 | Yes |
| 81 | Rapgef3 | 17766 | -0.378 | -0.1490 | Yes |
| 82 | Gp9 | 17768 | -0.378 | -0.1325 | Yes |
| 83 | Thbs1 | 18020 | -0.404 | -0.1279 | Yes |
| 84 | Adam9 | 18373 | -0.446 | -0.1267 | Yes |
| 85 | Itga2 | 18485 | -0.463 | -0.1122 | Yes |
| 86 | Ctso | 18535 | -0.469 | -0.0942 | Yes |
| 87 | Pecam1 | 18647 | -0.489 | -0.0786 | Yes |
| 88 | Hmgcs2 | 18720 | -0.506 | -0.0602 | Yes |
| 89 | Sirt2 | 18905 | -0.558 | -0.0453 | Yes |
| 90 | Olr1 | 19052 | -0.624 | -0.0256 | Yes |
| 91 | Iscu | 19277 | -0.887 | 0.0016 | Yes |