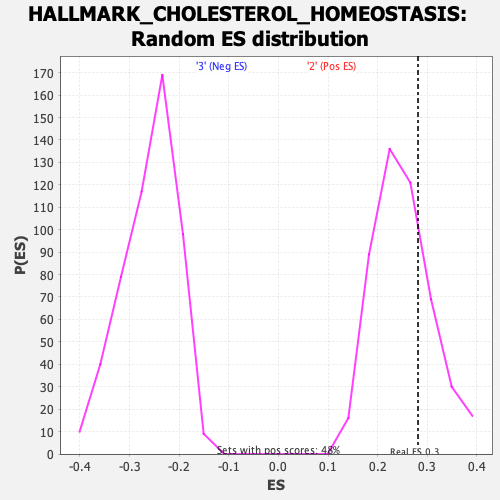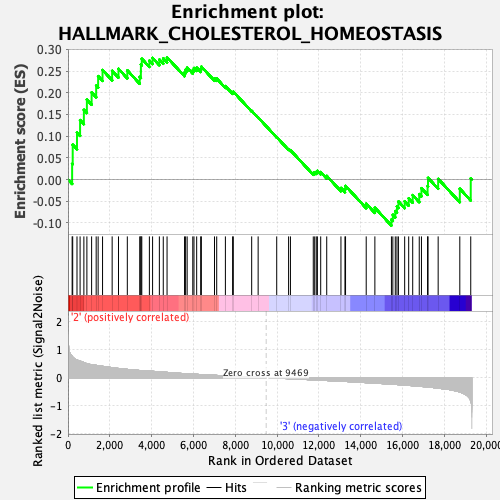
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.2813365 |
| Normalized Enrichment Score (NES) | 1.1252412 |
| Nominal p-value | 0.25104603 |
| FDR q-value | 0.9752792 |
| FWER p-Value | 0.96 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ldlr | 198 | 0.761 | 0.0362 | Yes |
| 2 | Trib3 | 225 | 0.741 | 0.0802 | Yes |
| 3 | Fdps | 430 | 0.632 | 0.1082 | Yes |
| 4 | Hmgcs1 | 578 | 0.595 | 0.1369 | Yes |
| 5 | Hmgcr | 759 | 0.541 | 0.1605 | Yes |
| 6 | Acat2 | 903 | 0.506 | 0.1840 | Yes |
| 7 | Nsdhl | 1129 | 0.465 | 0.2007 | Yes |
| 8 | Fdft1 | 1347 | 0.442 | 0.2165 | Yes |
| 9 | Hsd17b7 | 1441 | 0.434 | 0.2381 | Yes |
| 10 | Cyp51 | 1652 | 0.410 | 0.2523 | Yes |
| 11 | Pmvk | 2111 | 0.360 | 0.2505 | Yes |
| 12 | Trp53inp1 | 2414 | 0.332 | 0.2551 | Yes |
| 13 | Atxn2 | 2836 | 0.299 | 0.2514 | Yes |
| 14 | Sc5d | 3429 | 0.259 | 0.2365 | Yes |
| 15 | Gldc | 3484 | 0.255 | 0.2492 | Yes |
| 16 | Atf3 | 3486 | 0.255 | 0.2648 | Yes |
| 17 | Sqle | 3528 | 0.255 | 0.2782 | Yes |
| 18 | Gpx8 | 3891 | 0.239 | 0.2740 | Yes |
| 19 | Srebf2 | 4044 | 0.230 | 0.2801 | Yes |
| 20 | Ctnnb1 | 4368 | 0.213 | 0.2764 | Yes |
| 21 | Mvk | 4555 | 0.201 | 0.2790 | Yes |
| 22 | Pcyt2 | 4736 | 0.191 | 0.2813 | Yes |
| 23 | Niban1 | 5572 | 0.147 | 0.2469 | No |
| 24 | Lpl | 5623 | 0.145 | 0.2532 | No |
| 25 | Fasn | 5697 | 0.141 | 0.2580 | No |
| 26 | Lgals3 | 5966 | 0.129 | 0.2519 | No |
| 27 | Jag1 | 6023 | 0.128 | 0.2568 | No |
| 28 | Chka | 6152 | 0.122 | 0.2577 | No |
| 29 | Gnai1 | 6344 | 0.115 | 0.2547 | No |
| 30 | Nfil3 | 6376 | 0.113 | 0.2601 | No |
| 31 | Fabp5 | 7004 | 0.088 | 0.2328 | No |
| 32 | Abca2 | 7110 | 0.084 | 0.2325 | No |
| 33 | Alcam | 7525 | 0.070 | 0.2152 | No |
| 34 | Stard4 | 7870 | 0.056 | 0.2007 | No |
| 35 | Fads2 | 7903 | 0.054 | 0.2024 | No |
| 36 | Tmem97 | 8782 | 0.022 | 0.1581 | No |
| 37 | Cpeb2 | 9091 | 0.013 | 0.1429 | No |
| 38 | Pdk3 | 9979 | -0.012 | 0.0975 | No |
| 39 | Tnfrsf12a | 10547 | -0.031 | 0.0700 | No |
| 40 | Plscr1 | 10635 | -0.035 | 0.0676 | No |
| 41 | Ethe1 | 11727 | -0.075 | 0.0154 | No |
| 42 | Pparg | 11795 | -0.077 | 0.0167 | No |
| 43 | Tm7sf2 | 11870 | -0.080 | 0.0177 | No |
| 44 | Ebp | 11924 | -0.082 | 0.0200 | No |
| 45 | Gstm7 | 12082 | -0.087 | 0.0171 | No |
| 46 | S100a11 | 12370 | -0.099 | 0.0082 | No |
| 47 | Idi1 | 13050 | -0.123 | -0.0195 | No |
| 48 | Gusb | 13242 | -0.130 | -0.0215 | No |
| 49 | Errfi1 | 13270 | -0.132 | -0.0149 | No |
| 50 | Acss2 | 14258 | -0.174 | -0.0555 | No |
| 51 | Aldoc | 14672 | -0.194 | -0.0651 | No |
| 52 | Stx5a | 15464 | -0.230 | -0.0922 | No |
| 53 | Cd9 | 15531 | -0.233 | -0.0814 | No |
| 54 | Dhcr7 | 15648 | -0.239 | -0.0728 | No |
| 55 | Ech1 | 15728 | -0.242 | -0.0621 | No |
| 56 | Lss | 15798 | -0.246 | -0.0507 | No |
| 57 | Anxa5 | 16097 | -0.262 | -0.0502 | No |
| 58 | Lgmn | 16289 | -0.274 | -0.0434 | No |
| 59 | Atf5 | 16481 | -0.284 | -0.0360 | No |
| 60 | Pnrc1 | 16794 | -0.302 | -0.0337 | No |
| 61 | Plaur | 16899 | -0.311 | -0.0201 | No |
| 62 | Actg1 | 17192 | -0.327 | -0.0153 | No |
| 63 | Mvd | 17213 | -0.330 | 0.0038 | No |
| 64 | Clu | 17698 | -0.371 | 0.0013 | No |
| 65 | Antxr2 | 18733 | -0.509 | -0.0213 | No |
| 66 | Fbxo6 | 19256 | -0.837 | 0.0027 | No |