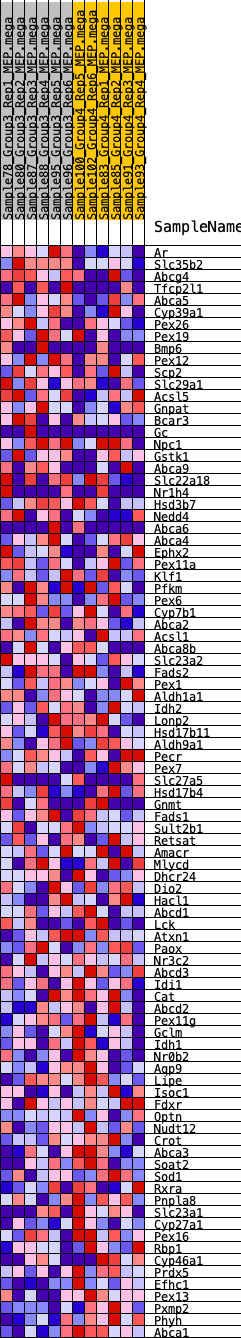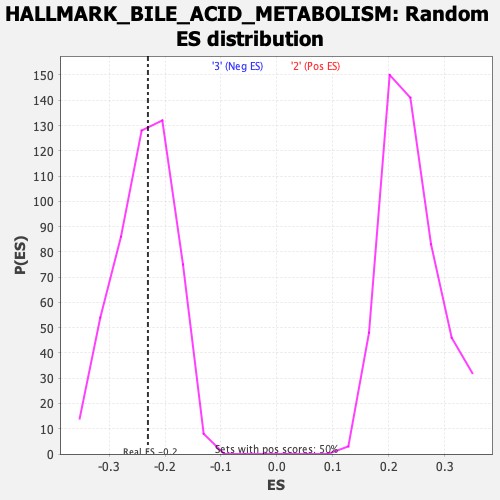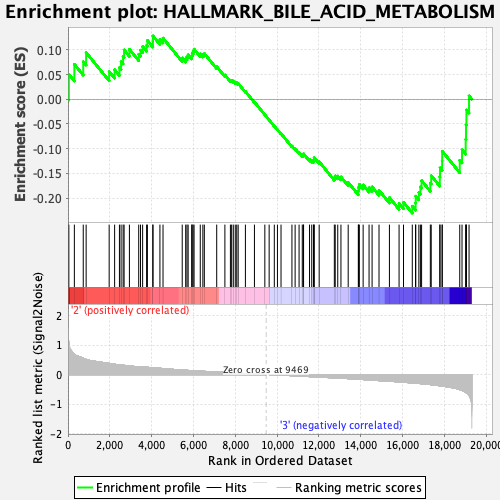
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.23057362 |
| Normalized Enrichment Score (NES) | -0.9725913 |
| Nominal p-value | 0.4889336 |
| FDR q-value | 0.93807745 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ar | 40 | 1.056 | 0.0505 | No |
| 2 | Slc35b2 | 304 | 0.684 | 0.0708 | No |
| 3 | Abcg4 | 729 | 0.552 | 0.0762 | No |
| 4 | Tfcp2l1 | 867 | 0.512 | 0.0945 | No |
| 5 | Abca5 | 1968 | 0.377 | 0.0560 | No |
| 6 | Cyp39a1 | 2229 | 0.348 | 0.0598 | No |
| 7 | Pex26 | 2459 | 0.328 | 0.0642 | No |
| 8 | Pex19 | 2538 | 0.323 | 0.0762 | No |
| 9 | Bmp6 | 2634 | 0.314 | 0.0869 | No |
| 10 | Pex12 | 2682 | 0.311 | 0.0999 | No |
| 11 | Scp2 | 2936 | 0.291 | 0.1012 | No |
| 12 | Slc29a1 | 3381 | 0.262 | 0.0911 | No |
| 13 | Acsl5 | 3467 | 0.255 | 0.0994 | No |
| 14 | Gnpat | 3568 | 0.252 | 0.1067 | No |
| 15 | Bcar3 | 3758 | 0.244 | 0.1090 | No |
| 16 | Gc | 3796 | 0.243 | 0.1191 | No |
| 17 | Npc1 | 4058 | 0.229 | 0.1170 | No |
| 18 | Gstk1 | 4060 | 0.229 | 0.1283 | No |
| 19 | Abca9 | 4395 | 0.212 | 0.1214 | No |
| 20 | Slc22a18 | 4543 | 0.202 | 0.1238 | No |
| 21 | Nr1h4 | 5461 | 0.152 | 0.0837 | No |
| 22 | Hsd3b7 | 5627 | 0.144 | 0.0823 | No |
| 23 | Nedd4 | 5688 | 0.141 | 0.0862 | No |
| 24 | Abca6 | 5744 | 0.139 | 0.0902 | No |
| 25 | Abca4 | 5914 | 0.131 | 0.0879 | No |
| 26 | Ephx2 | 5936 | 0.130 | 0.0933 | No |
| 27 | Pex11a | 5981 | 0.129 | 0.0974 | No |
| 28 | Klf1 | 6027 | 0.128 | 0.1014 | No |
| 29 | Pfkm | 6324 | 0.115 | 0.0918 | No |
| 30 | Pex6 | 6448 | 0.110 | 0.0909 | No |
| 31 | Cyp7b1 | 6518 | 0.107 | 0.0926 | No |
| 32 | Abca2 | 7110 | 0.084 | 0.0660 | No |
| 33 | Acsl1 | 7501 | 0.071 | 0.0493 | No |
| 34 | Abca8b | 7765 | 0.059 | 0.0385 | No |
| 35 | Slc23a2 | 7824 | 0.057 | 0.0383 | No |
| 36 | Fads2 | 7903 | 0.054 | 0.0370 | No |
| 37 | Pex1 | 7997 | 0.050 | 0.0346 | No |
| 38 | Aldh1a1 | 8068 | 0.048 | 0.0334 | No |
| 39 | Idh2 | 8136 | 0.045 | 0.0321 | No |
| 40 | Lonp2 | 8482 | 0.033 | 0.0158 | No |
| 41 | Hsd17b11 | 8914 | 0.019 | -0.0057 | No |
| 42 | Aldh9a1 | 9411 | 0.002 | -0.0314 | No |
| 43 | Pecr | 9617 | -0.000 | -0.0420 | No |
| 44 | Pex7 | 9864 | -0.008 | -0.0544 | No |
| 45 | Slc27a5 | 10014 | -0.013 | -0.0615 | No |
| 46 | Hsd17b4 | 10185 | -0.019 | -0.0694 | No |
| 47 | Gnmt | 10707 | -0.037 | -0.0947 | No |
| 48 | Fads1 | 10859 | -0.042 | -0.1005 | No |
| 49 | Sult2b1 | 11048 | -0.049 | -0.1078 | No |
| 50 | Retsat | 11201 | -0.055 | -0.1130 | No |
| 51 | Amacr | 11252 | -0.057 | -0.1127 | No |
| 52 | Mlycd | 11263 | -0.057 | -0.1104 | No |
| 53 | Dhcr24 | 11548 | -0.068 | -0.1218 | No |
| 54 | Dio2 | 11654 | -0.072 | -0.1237 | No |
| 55 | Hacl1 | 11737 | -0.075 | -0.1242 | No |
| 56 | Abcd1 | 11765 | -0.076 | -0.1218 | No |
| 57 | Lck | 11767 | -0.076 | -0.1181 | No |
| 58 | Atxn1 | 12009 | -0.084 | -0.1264 | No |
| 59 | Paox | 12734 | -0.112 | -0.1586 | No |
| 60 | Nr3c2 | 12776 | -0.114 | -0.1550 | No |
| 61 | Abcd3 | 12897 | -0.118 | -0.1554 | No |
| 62 | Idi1 | 13050 | -0.123 | -0.1572 | No |
| 63 | Cat | 13396 | -0.137 | -0.1683 | No |
| 64 | Abcd2 | 13881 | -0.157 | -0.1857 | No |
| 65 | Pex11g | 13891 | -0.158 | -0.1783 | No |
| 66 | Gclm | 13930 | -0.159 | -0.1724 | No |
| 67 | Idh1 | 14111 | -0.168 | -0.1734 | No |
| 68 | Nr0b2 | 14394 | -0.180 | -0.1791 | No |
| 69 | Aqp9 | 14539 | -0.187 | -0.1773 | No |
| 70 | Lipe | 14870 | -0.204 | -0.1843 | No |
| 71 | Isoc1 | 15368 | -0.225 | -0.1990 | No |
| 72 | Fdxr | 15829 | -0.247 | -0.2106 | No |
| 73 | Optn | 16041 | -0.258 | -0.2088 | No |
| 74 | Nudt12 | 16461 | -0.283 | -0.2165 | Yes |
| 75 | Crot | 16617 | -0.291 | -0.2101 | Yes |
| 76 | Abca3 | 16627 | -0.292 | -0.1961 | Yes |
| 77 | Soat2 | 16775 | -0.301 | -0.1887 | Yes |
| 78 | Sod1 | 16849 | -0.306 | -0.1773 | Yes |
| 79 | Rxra | 16906 | -0.311 | -0.1648 | Yes |
| 80 | Pnpla8 | 17324 | -0.339 | -0.1696 | Yes |
| 81 | Slc23a1 | 17366 | -0.343 | -0.1547 | Yes |
| 82 | Cyp27a1 | 17776 | -0.379 | -0.1571 | Yes |
| 83 | Pex16 | 17788 | -0.381 | -0.1387 | Yes |
| 84 | Rbp1 | 17889 | -0.390 | -0.1245 | Yes |
| 85 | Cyp46a1 | 17892 | -0.391 | -0.1052 | Yes |
| 86 | Prdx5 | 18730 | -0.508 | -0.1235 | Yes |
| 87 | Efhc1 | 18839 | -0.540 | -0.1022 | Yes |
| 88 | Pex13 | 19014 | -0.604 | -0.0813 | Yes |
| 89 | Pxmp2 | 19031 | -0.612 | -0.0517 | Yes |
| 90 | Phyh | 19051 | -0.623 | -0.0217 | Yes |
| 91 | Abca1 | 19175 | -0.703 | 0.0069 | Yes |