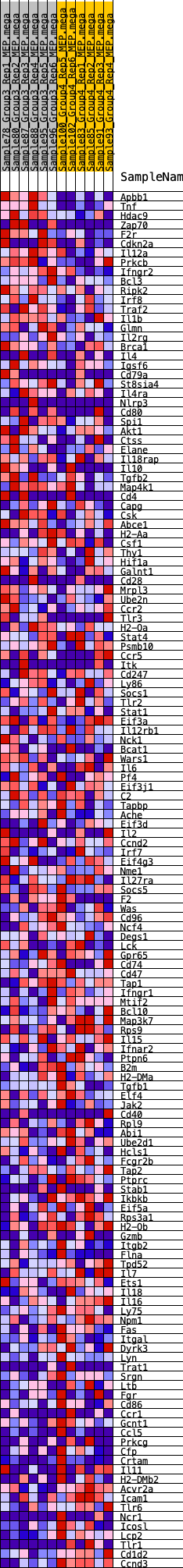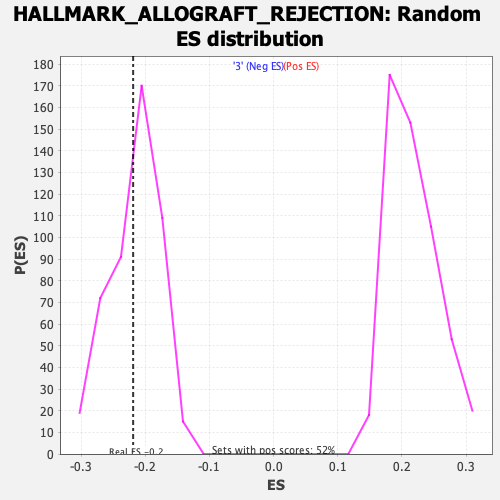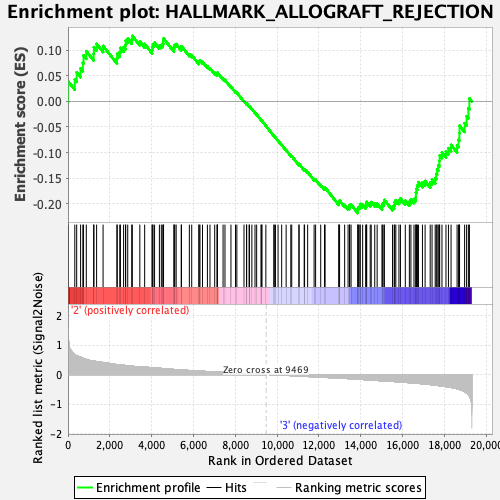
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group3_versus_Group4.MEP.mega_Pheno.cls #Group3_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group3_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ALLOGRAFT_REJECTION |
| Enrichment Score (ES) | -0.21880896 |
| Normalized Enrichment Score (NES) | -1.0109137 |
| Nominal p-value | 0.3970588 |
| FDR q-value | 0.8976989 |
| FWER p-Value | 0.996 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Apbb1 | 8 | 1.347 | 0.0395 | No |
| 2 | Tnf | 325 | 0.675 | 0.0430 | No |
| 3 | Hdac9 | 415 | 0.635 | 0.0571 | No |
| 4 | Zap70 | 602 | 0.589 | 0.0649 | No |
| 5 | F2r | 711 | 0.558 | 0.0757 | No |
| 6 | Cdkn2a | 743 | 0.548 | 0.0904 | No |
| 7 | Il12a | 875 | 0.510 | 0.0986 | No |
| 8 | Prkcb | 1226 | 0.453 | 0.0938 | No |
| 9 | Ifngr2 | 1240 | 0.452 | 0.1065 | No |
| 10 | Bcl3 | 1368 | 0.441 | 0.1129 | No |
| 11 | Ripk2 | 1677 | 0.406 | 0.1089 | No |
| 12 | Irf8 | 2339 | 0.337 | 0.0844 | No |
| 13 | Traf2 | 2353 | 0.336 | 0.0936 | No |
| 14 | Il1b | 2476 | 0.327 | 0.0970 | No |
| 15 | Glmn | 2506 | 0.325 | 0.1051 | No |
| 16 | Il2rg | 2658 | 0.313 | 0.1064 | No |
| 17 | Brca1 | 2749 | 0.306 | 0.1108 | No |
| 18 | Il4 | 2760 | 0.305 | 0.1193 | No |
| 19 | Igsf6 | 2856 | 0.297 | 0.1232 | No |
| 20 | Cd79a | 3047 | 0.282 | 0.1216 | No |
| 21 | St8sia4 | 3074 | 0.281 | 0.1286 | No |
| 22 | Il4ra | 3434 | 0.258 | 0.1175 | No |
| 23 | Nlrp3 | 3665 | 0.246 | 0.1128 | No |
| 24 | Cd80 | 4020 | 0.232 | 0.1012 | No |
| 25 | Spi1 | 4030 | 0.231 | 0.1075 | No |
| 26 | Akt1 | 4066 | 0.229 | 0.1125 | No |
| 27 | Ctss | 4139 | 0.224 | 0.1154 | No |
| 28 | Elane | 4370 | 0.213 | 0.1097 | No |
| 29 | Il18rap | 4462 | 0.207 | 0.1111 | No |
| 30 | Il10 | 4535 | 0.203 | 0.1133 | No |
| 31 | Tgfb2 | 4554 | 0.201 | 0.1183 | No |
| 32 | Map4k1 | 4568 | 0.201 | 0.1236 | No |
| 33 | Cd4 | 5066 | 0.172 | 0.1028 | No |
| 34 | Capg | 5073 | 0.172 | 0.1075 | No |
| 35 | Csk | 5106 | 0.170 | 0.1109 | No |
| 36 | Abce1 | 5171 | 0.166 | 0.1125 | No |
| 37 | H2-Aa | 5413 | 0.154 | 0.1045 | No |
| 38 | Csf1 | 5426 | 0.153 | 0.1084 | No |
| 39 | Thy1 | 5802 | 0.136 | 0.0928 | No |
| 40 | Hif1a | 5915 | 0.131 | 0.0909 | No |
| 41 | Galnt1 | 6254 | 0.117 | 0.0767 | No |
| 42 | Cd28 | 6271 | 0.117 | 0.0793 | No |
| 43 | Mrpl3 | 6310 | 0.116 | 0.0808 | No |
| 44 | Ube2n | 6431 | 0.111 | 0.0778 | No |
| 45 | Ccr2 | 6665 | 0.101 | 0.0686 | No |
| 46 | Tlr3 | 6788 | 0.097 | 0.0651 | No |
| 47 | H2-Oa | 7014 | 0.087 | 0.0560 | No |
| 48 | Stat4 | 7124 | 0.083 | 0.0528 | No |
| 49 | Psmb10 | 7144 | 0.082 | 0.0542 | No |
| 50 | Ccr5 | 7145 | 0.082 | 0.0566 | No |
| 51 | Itk | 7414 | 0.073 | 0.0448 | No |
| 52 | Cd247 | 7505 | 0.071 | 0.0422 | No |
| 53 | Ly86 | 7791 | 0.058 | 0.0290 | No |
| 54 | Socs1 | 8008 | 0.050 | 0.0193 | No |
| 55 | Tlr2 | 8073 | 0.048 | 0.0173 | No |
| 56 | Stat1 | 8415 | 0.035 | 0.0006 | No |
| 57 | Eif3a | 8536 | 0.031 | -0.0048 | No |
| 58 | Il12rb1 | 8542 | 0.030 | -0.0041 | No |
| 59 | Nck1 | 8661 | 0.027 | -0.0095 | No |
| 60 | Bcat1 | 8673 | 0.027 | -0.0093 | No |
| 61 | Wars1 | 8791 | 0.022 | -0.0147 | No |
| 62 | Il6 | 8941 | 0.018 | -0.0220 | No |
| 63 | Pf4 | 9020 | 0.015 | -0.0256 | No |
| 64 | Eif3j1 | 9227 | 0.008 | -0.0361 | No |
| 65 | C2 | 9265 | 0.007 | -0.0378 | No |
| 66 | Tapbp | 9267 | 0.007 | -0.0377 | No |
| 67 | Ache | 9460 | 0.000 | -0.0477 | No |
| 68 | Eif3d | 9839 | -0.007 | -0.0672 | No |
| 69 | Il2 | 9863 | -0.008 | -0.0681 | No |
| 70 | Ccnd2 | 9919 | -0.010 | -0.0707 | No |
| 71 | Irf7 | 10043 | -0.014 | -0.0767 | No |
| 72 | Eif4g3 | 10216 | -0.020 | -0.0851 | No |
| 73 | Nme1 | 10430 | -0.028 | -0.0954 | No |
| 74 | Il27ra | 10653 | -0.035 | -0.1059 | No |
| 75 | Socs5 | 10697 | -0.037 | -0.1071 | No |
| 76 | F2 | 11044 | -0.049 | -0.1237 | No |
| 77 | Was | 11045 | -0.049 | -0.1222 | No |
| 78 | Cd96 | 11292 | -0.058 | -0.1333 | No |
| 79 | Ncf4 | 11311 | -0.059 | -0.1325 | No |
| 80 | Degs1 | 11458 | -0.065 | -0.1382 | No |
| 81 | Lck | 11767 | -0.076 | -0.1521 | No |
| 82 | Gpr65 | 11838 | -0.079 | -0.1534 | No |
| 83 | Cd74 | 12083 | -0.087 | -0.1635 | No |
| 84 | Cd47 | 12264 | -0.094 | -0.1702 | No |
| 85 | Tap1 | 12292 | -0.095 | -0.1688 | No |
| 86 | Ifngr1 | 12945 | -0.120 | -0.1992 | No |
| 87 | Mtif2 | 12963 | -0.120 | -0.1966 | No |
| 88 | Bcl10 | 12985 | -0.121 | -0.1941 | No |
| 89 | Map3k7 | 13227 | -0.130 | -0.2028 | No |
| 90 | Rps9 | 13391 | -0.137 | -0.2073 | No |
| 91 | Il15 | 13461 | -0.140 | -0.2067 | No |
| 92 | Ifnar2 | 13467 | -0.140 | -0.2028 | No |
| 93 | Ptpn6 | 13534 | -0.143 | -0.2021 | No |
| 94 | B2m | 13856 | -0.156 | -0.2142 | Yes |
| 95 | H2-DMa | 13860 | -0.156 | -0.2097 | Yes |
| 96 | Tgfb1 | 13896 | -0.158 | -0.2069 | Yes |
| 97 | Elf4 | 13963 | -0.161 | -0.2056 | Yes |
| 98 | Jak2 | 13966 | -0.161 | -0.2009 | Yes |
| 99 | Cd40 | 14088 | -0.167 | -0.2023 | Yes |
| 100 | Rpl9 | 14232 | -0.173 | -0.2046 | Yes |
| 101 | Abi1 | 14268 | -0.175 | -0.2013 | Yes |
| 102 | Ube2d1 | 14282 | -0.175 | -0.1967 | Yes |
| 103 | Hcls1 | 14449 | -0.183 | -0.2000 | Yes |
| 104 | Fcgr2b | 14497 | -0.185 | -0.1970 | Yes |
| 105 | Tap2 | 14666 | -0.194 | -0.2000 | Yes |
| 106 | Ptprc | 14781 | -0.200 | -0.2000 | Yes |
| 107 | Stab1 | 15017 | -0.207 | -0.2061 | Yes |
| 108 | Ikbkb | 15026 | -0.208 | -0.2004 | Yes |
| 109 | Eif5a | 15104 | -0.212 | -0.1982 | Yes |
| 110 | Rps3a1 | 15126 | -0.213 | -0.1929 | Yes |
| 111 | H2-Ob | 15516 | -0.232 | -0.2064 | Yes |
| 112 | Gzmb | 15595 | -0.237 | -0.2034 | Yes |
| 113 | Itgb2 | 15619 | -0.238 | -0.1976 | Yes |
| 114 | Flna | 15674 | -0.240 | -0.1933 | Yes |
| 115 | Tpd52 | 15823 | -0.247 | -0.1937 | Yes |
| 116 | Il7 | 15895 | -0.252 | -0.1900 | Yes |
| 117 | Ets1 | 16127 | -0.264 | -0.1942 | Yes |
| 118 | Il18 | 16321 | -0.274 | -0.1961 | Yes |
| 119 | Il16 | 16395 | -0.279 | -0.1917 | Yes |
| 120 | Ly75 | 16543 | -0.287 | -0.1908 | Yes |
| 121 | Npm1 | 16632 | -0.292 | -0.1868 | Yes |
| 122 | Fas | 16633 | -0.292 | -0.1781 | Yes |
| 123 | Itgal | 16664 | -0.294 | -0.1710 | Yes |
| 124 | Dyrk3 | 16707 | -0.297 | -0.1644 | Yes |
| 125 | Lyn | 16756 | -0.300 | -0.1580 | Yes |
| 126 | Trat1 | 16947 | -0.314 | -0.1586 | Yes |
| 127 | Srgn | 17074 | -0.321 | -0.1557 | Yes |
| 128 | Ltb | 17317 | -0.338 | -0.1583 | Yes |
| 129 | Fgr | 17406 | -0.346 | -0.1526 | Yes |
| 130 | Cd86 | 17556 | -0.360 | -0.1498 | Yes |
| 131 | Ccr1 | 17608 | -0.362 | -0.1417 | Yes |
| 132 | Gcnt1 | 17648 | -0.366 | -0.1329 | Yes |
| 133 | Ccl5 | 17704 | -0.372 | -0.1248 | Yes |
| 134 | Prkcg | 17759 | -0.378 | -0.1164 | Yes |
| 135 | Cfp | 17770 | -0.379 | -0.1057 | Yes |
| 136 | Crtam | 17880 | -0.390 | -0.0998 | Yes |
| 137 | Il11 | 18072 | -0.409 | -0.0977 | Yes |
| 138 | H2-DMb2 | 18190 | -0.417 | -0.0914 | Yes |
| 139 | Acvr2a | 18315 | -0.436 | -0.0850 | Yes |
| 140 | Icam1 | 18603 | -0.480 | -0.0857 | Yes |
| 141 | Tlr6 | 18674 | -0.493 | -0.0748 | Yes |
| 142 | Ncr1 | 18714 | -0.505 | -0.0619 | Yes |
| 143 | Icosl | 18721 | -0.506 | -0.0472 | Yes |
| 144 | Lcp2 | 18963 | -0.584 | -0.0425 | Yes |
| 145 | Tlr1 | 19058 | -0.624 | -0.0289 | Yes |
| 146 | Cd1d2 | 19143 | -0.680 | -0.0131 | Yes |
| 147 | Ccnd3 | 19192 | -0.729 | 0.0060 | Yes |