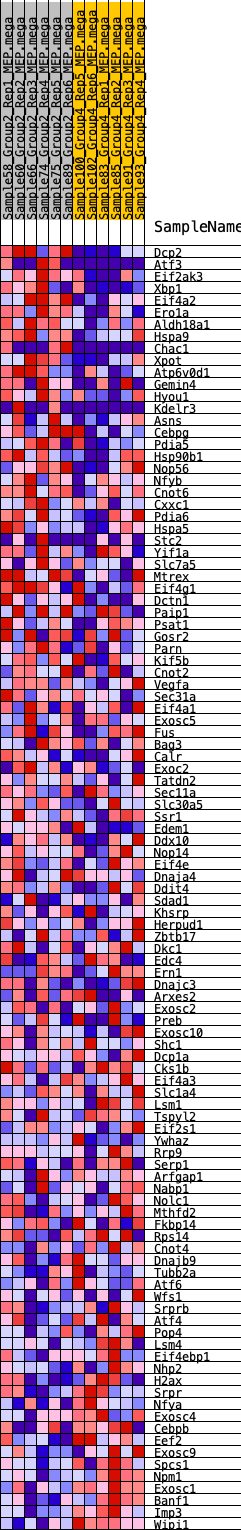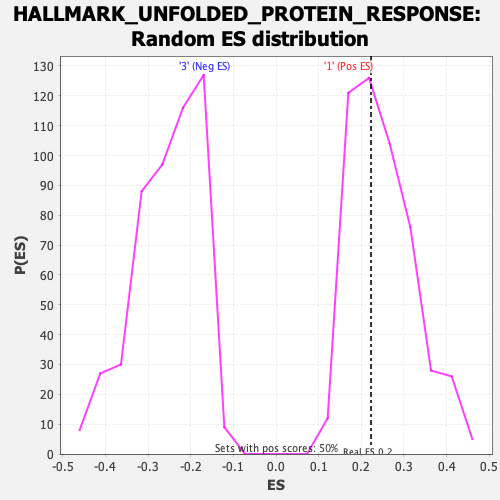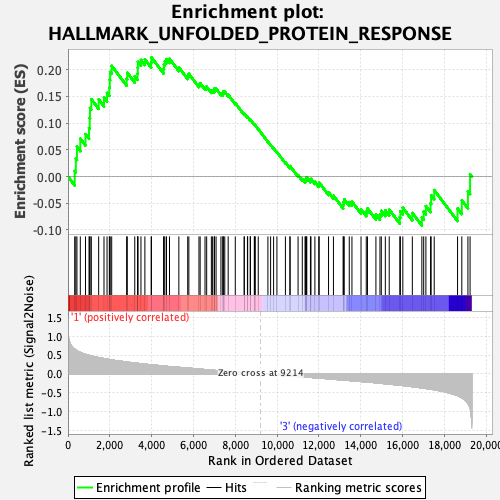
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.22288616 |
| Normalized Enrichment Score (NES) | 0.89192414 |
| Nominal p-value | 0.5763052 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.999 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 320 | 0.657 | 0.0103 | Yes |
| 2 | Atf3 | 374 | 0.633 | 0.0335 | Yes |
| 3 | Eif2ak3 | 427 | 0.610 | 0.0559 | Yes |
| 4 | Xbp1 | 588 | 0.570 | 0.0710 | Yes |
| 5 | Eif4a2 | 836 | 0.519 | 0.0794 | Yes |
| 6 | Ero1a | 1006 | 0.492 | 0.0908 | Yes |
| 7 | Aldh18a1 | 1037 | 0.488 | 0.1093 | Yes |
| 8 | Hspa9 | 1053 | 0.485 | 0.1284 | Yes |
| 9 | Chac1 | 1115 | 0.477 | 0.1448 | Yes |
| 10 | Xpot | 1466 | 0.434 | 0.1444 | Yes |
| 11 | Atp6v0d1 | 1721 | 0.408 | 0.1479 | Yes |
| 12 | Gemin4 | 1866 | 0.394 | 0.1566 | Yes |
| 13 | Hyou1 | 1976 | 0.381 | 0.1665 | Yes |
| 14 | Kdelr3 | 1997 | 0.379 | 0.1811 | Yes |
| 15 | Asns | 2014 | 0.378 | 0.1958 | Yes |
| 16 | Cebpg | 2089 | 0.371 | 0.2071 | Yes |
| 17 | Pdia5 | 2803 | 0.316 | 0.1830 | Yes |
| 18 | Hsp90b1 | 2834 | 0.313 | 0.1943 | Yes |
| 19 | Nop56 | 3192 | 0.291 | 0.1876 | Yes |
| 20 | Nfyb | 3325 | 0.283 | 0.1924 | Yes |
| 21 | Cnot6 | 3335 | 0.282 | 0.2035 | Yes |
| 22 | Cxxc1 | 3343 | 0.282 | 0.2147 | Yes |
| 23 | Pdia6 | 3488 | 0.271 | 0.2183 | Yes |
| 24 | Hspa5 | 3676 | 0.260 | 0.2193 | Yes |
| 25 | Stc2 | 3976 | 0.243 | 0.2137 | Yes |
| 26 | Yif1a | 3991 | 0.242 | 0.2229 | Yes |
| 27 | Slc7a5 | 4571 | 0.212 | 0.2014 | No |
| 28 | Mtrex | 4586 | 0.210 | 0.2093 | No |
| 29 | Eif4g1 | 4631 | 0.208 | 0.2156 | No |
| 30 | Dctn1 | 4710 | 0.202 | 0.2198 | No |
| 31 | Paip1 | 4853 | 0.194 | 0.2204 | No |
| 32 | Psat1 | 5300 | 0.175 | 0.2043 | No |
| 33 | Gosr2 | 5724 | 0.157 | 0.1887 | No |
| 34 | Parn | 5778 | 0.154 | 0.1923 | No |
| 35 | Kif5b | 6261 | 0.129 | 0.1725 | No |
| 36 | Cnot2 | 6318 | 0.127 | 0.1748 | No |
| 37 | Vegfa | 6551 | 0.116 | 0.1674 | No |
| 38 | Sec31a | 6630 | 0.112 | 0.1680 | No |
| 39 | Eif4a1 | 6854 | 0.101 | 0.1605 | No |
| 40 | Exosc5 | 6914 | 0.098 | 0.1614 | No |
| 41 | Fus | 6994 | 0.094 | 0.1612 | No |
| 42 | Bag3 | 7000 | 0.094 | 0.1648 | No |
| 43 | Calr | 7079 | 0.090 | 0.1645 | No |
| 44 | Exoc2 | 7313 | 0.081 | 0.1557 | No |
| 45 | Tatdn2 | 7403 | 0.076 | 0.1542 | No |
| 46 | Sec11a | 7419 | 0.076 | 0.1565 | No |
| 47 | Slc30a5 | 7422 | 0.076 | 0.1595 | No |
| 48 | Ssr1 | 7495 | 0.072 | 0.1587 | No |
| 49 | Edem1 | 7658 | 0.066 | 0.1529 | No |
| 50 | Ddx10 | 7998 | 0.052 | 0.1374 | No |
| 51 | Nop14 | 8424 | 0.033 | 0.1166 | No |
| 52 | Eif4e | 8428 | 0.033 | 0.1178 | No |
| 53 | Dnaja4 | 8579 | 0.025 | 0.1110 | No |
| 54 | Ddit4 | 8590 | 0.025 | 0.1115 | No |
| 55 | Sdad1 | 8717 | 0.019 | 0.1057 | No |
| 56 | Khsrp | 8718 | 0.019 | 0.1065 | No |
| 57 | Herpud1 | 8911 | 0.013 | 0.0970 | No |
| 58 | Zbtb17 | 8913 | 0.013 | 0.0975 | No |
| 59 | Dkc1 | 8955 | 0.011 | 0.0958 | No |
| 60 | Edc4 | 9096 | 0.005 | 0.0888 | No |
| 61 | Ern1 | 9558 | -0.011 | 0.0652 | No |
| 62 | Dnajc3 | 9687 | -0.017 | 0.0592 | No |
| 63 | Arxes2 | 9830 | -0.023 | 0.0528 | No |
| 64 | Exosc2 | 9989 | -0.030 | 0.0458 | No |
| 65 | Preb | 10395 | -0.046 | 0.0266 | No |
| 66 | Exosc10 | 10613 | -0.055 | 0.0175 | No |
| 67 | Shc1 | 10616 | -0.055 | 0.0197 | No |
| 68 | Dcp1a | 11000 | -0.070 | 0.0026 | No |
| 69 | Cks1b | 11202 | -0.078 | -0.0047 | No |
| 70 | Eif4a3 | 11336 | -0.084 | -0.0081 | No |
| 71 | Slc1a4 | 11351 | -0.084 | -0.0054 | No |
| 72 | Lsm1 | 11408 | -0.087 | -0.0048 | No |
| 73 | Tspyl2 | 11413 | -0.087 | -0.0014 | No |
| 74 | Eif2s1 | 11592 | -0.092 | -0.0069 | No |
| 75 | Ywhaz | 11617 | -0.093 | -0.0043 | No |
| 76 | Rrp9 | 11804 | -0.102 | -0.0098 | No |
| 77 | Serp1 | 11990 | -0.110 | -0.0150 | No |
| 78 | Arfgap1 | 12008 | -0.111 | -0.0113 | No |
| 79 | Nabp1 | 12457 | -0.130 | -0.0293 | No |
| 80 | Nolc1 | 12691 | -0.140 | -0.0357 | No |
| 81 | Mthfd2 | 13153 | -0.161 | -0.0531 | No |
| 82 | Fkbp14 | 13176 | -0.162 | -0.0476 | No |
| 83 | Rps14 | 13206 | -0.164 | -0.0424 | No |
| 84 | Cnot4 | 13448 | -0.175 | -0.0477 | No |
| 85 | Dnajb9 | 13576 | -0.181 | -0.0469 | No |
| 86 | Tubb2a | 14010 | -0.202 | -0.0612 | No |
| 87 | Atf6 | 14260 | -0.212 | -0.0655 | No |
| 88 | Wfs1 | 14318 | -0.214 | -0.0596 | No |
| 89 | Srprb | 14722 | -0.235 | -0.0710 | No |
| 90 | Atf4 | 14916 | -0.246 | -0.0709 | No |
| 91 | Pop4 | 14980 | -0.249 | -0.0640 | No |
| 92 | Lsm4 | 15172 | -0.259 | -0.0633 | No |
| 93 | Eif4ebp1 | 15358 | -0.270 | -0.0618 | No |
| 94 | Nhp2 | 15865 | -0.299 | -0.0759 | No |
| 95 | H2ax | 15892 | -0.301 | -0.0649 | No |
| 96 | Srpr | 16009 | -0.308 | -0.0584 | No |
| 97 | Nfya | 16463 | -0.338 | -0.0681 | No |
| 98 | Exosc4 | 16919 | -0.371 | -0.0765 | No |
| 99 | Cebpb | 17006 | -0.378 | -0.0655 | No |
| 100 | Eef2 | 17112 | -0.386 | -0.0551 | No |
| 101 | Exosc9 | 17334 | -0.403 | -0.0501 | No |
| 102 | Spcs1 | 17362 | -0.405 | -0.0349 | No |
| 103 | Npm1 | 17509 | -0.418 | -0.0253 | No |
| 104 | Exosc1 | 18625 | -0.576 | -0.0597 | No |
| 105 | Banf1 | 18829 | -0.630 | -0.0444 | No |
| 106 | Imp3 | 19121 | -0.778 | -0.0276 | No |
| 107 | Wipi1 | 19224 | -0.907 | 0.0043 | No |