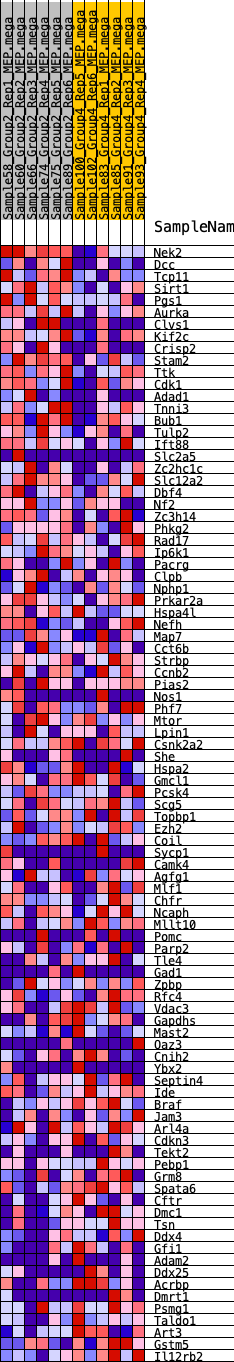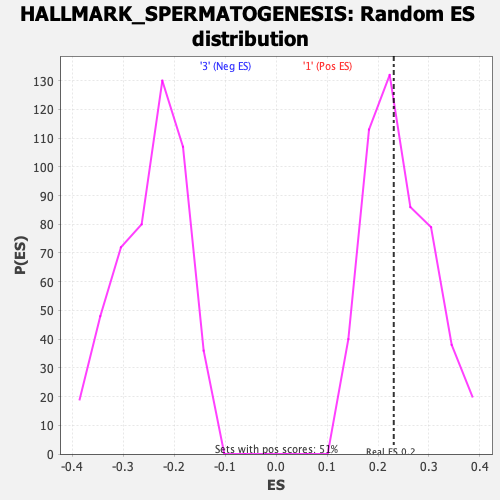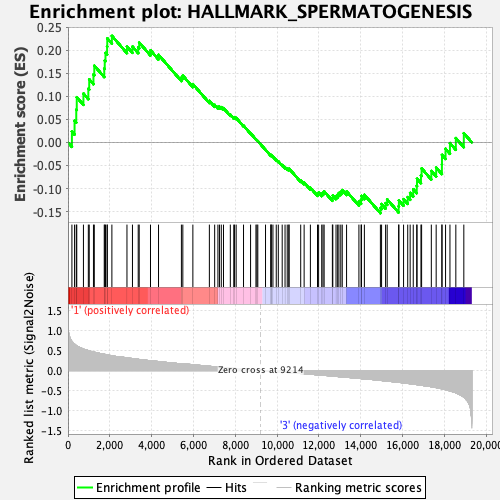
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.2308183 |
| Normalized Enrichment Score (NES) | 0.95199937 |
| Nominal p-value | 0.507874 |
| FDR q-value | 0.9872198 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nek2 | 187 | 0.727 | 0.0234 | Yes |
| 2 | Dcc | 314 | 0.658 | 0.0469 | Yes |
| 3 | Tcp11 | 395 | 0.621 | 0.0711 | Yes |
| 4 | Sirt1 | 419 | 0.614 | 0.0979 | Yes |
| 5 | Pgs1 | 738 | 0.537 | 0.1058 | Yes |
| 6 | Aurka | 972 | 0.498 | 0.1164 | Yes |
| 7 | Clvs1 | 1015 | 0.491 | 0.1366 | Yes |
| 8 | Kif2c | 1217 | 0.465 | 0.1473 | Yes |
| 9 | Crisp2 | 1257 | 0.460 | 0.1663 | Yes |
| 10 | Stam2 | 1733 | 0.407 | 0.1601 | Yes |
| 11 | Ttk | 1757 | 0.405 | 0.1774 | Yes |
| 12 | Cdk1 | 1792 | 0.400 | 0.1938 | Yes |
| 13 | Adad1 | 1869 | 0.394 | 0.2078 | Yes |
| 14 | Tnni3 | 1878 | 0.393 | 0.2253 | Yes |
| 15 | Bub1 | 2098 | 0.370 | 0.2308 | Yes |
| 16 | Tulp2 | 2819 | 0.314 | 0.2077 | No |
| 17 | Ift88 | 3082 | 0.299 | 0.2077 | No |
| 18 | Slc2a5 | 3357 | 0.280 | 0.2062 | No |
| 19 | Zc2hc1c | 3404 | 0.276 | 0.2164 | No |
| 20 | Slc12a2 | 3944 | 0.245 | 0.1995 | No |
| 21 | Dbf4 | 4328 | 0.223 | 0.1898 | No |
| 22 | Nf2 | 5422 | 0.169 | 0.1406 | No |
| 23 | Zc3h14 | 5490 | 0.165 | 0.1446 | No |
| 24 | Phkg2 | 5970 | 0.144 | 0.1263 | No |
| 25 | Rad17 | 6759 | 0.106 | 0.0901 | No |
| 26 | Ip6k1 | 7012 | 0.094 | 0.0812 | No |
| 27 | Pacrg | 7168 | 0.087 | 0.0771 | No |
| 28 | Clpb | 7241 | 0.083 | 0.0772 | No |
| 29 | Nphp1 | 7329 | 0.080 | 0.0763 | No |
| 30 | Prkar2a | 7434 | 0.075 | 0.0743 | No |
| 31 | Hspa4l | 7761 | 0.061 | 0.0601 | No |
| 32 | Nefh | 7926 | 0.055 | 0.0541 | No |
| 33 | Map7 | 7964 | 0.053 | 0.0546 | No |
| 34 | Cct6b | 8040 | 0.050 | 0.0530 | No |
| 35 | Strbp | 8392 | 0.035 | 0.0363 | No |
| 36 | Ccnb2 | 8729 | 0.019 | 0.0197 | No |
| 37 | Pias2 | 8986 | 0.010 | 0.0068 | No |
| 38 | Nos1 | 9032 | 0.008 | 0.0048 | No |
| 39 | Phf7 | 9075 | 0.006 | 0.0029 | No |
| 40 | Mtor | 9444 | -0.007 | -0.0160 | No |
| 41 | Lpin1 | 9703 | -0.018 | -0.0286 | No |
| 42 | Csnk2a2 | 9708 | -0.018 | -0.0280 | No |
| 43 | She | 9712 | -0.018 | -0.0273 | No |
| 44 | Hspa2 | 9798 | -0.021 | -0.0307 | No |
| 45 | Gmcl1 | 9957 | -0.029 | -0.0377 | No |
| 46 | Pcsk4 | 10055 | -0.032 | -0.0413 | No |
| 47 | Scg5 | 10241 | -0.040 | -0.0490 | No |
| 48 | Topbp1 | 10377 | -0.046 | -0.0540 | No |
| 49 | Ezh2 | 10482 | -0.050 | -0.0571 | No |
| 50 | Coil | 10557 | -0.053 | -0.0586 | No |
| 51 | Sycp1 | 10571 | -0.053 | -0.0568 | No |
| 52 | Camk4 | 11126 | -0.075 | -0.0822 | No |
| 53 | Agfg1 | 11290 | -0.082 | -0.0870 | No |
| 54 | Mlf1 | 11589 | -0.092 | -0.0983 | No |
| 55 | Chfr | 11929 | -0.108 | -0.1110 | No |
| 56 | Ncaph | 11982 | -0.110 | -0.1087 | No |
| 57 | Mllt10 | 12131 | -0.116 | -0.1111 | No |
| 58 | Pomc | 12191 | -0.118 | -0.1088 | No |
| 59 | Parp2 | 12246 | -0.120 | -0.1061 | No |
| 60 | Tle4 | 12640 | -0.138 | -0.1203 | No |
| 61 | Gad1 | 12661 | -0.139 | -0.1150 | No |
| 62 | Zpbp | 12815 | -0.146 | -0.1163 | No |
| 63 | Rfc4 | 12893 | -0.150 | -0.1134 | No |
| 64 | Vdac3 | 12953 | -0.153 | -0.1095 | No |
| 65 | Gapdhs | 13043 | -0.156 | -0.1070 | No |
| 66 | Mast2 | 13112 | -0.159 | -0.1033 | No |
| 67 | Oaz3 | 13319 | -0.169 | -0.1063 | No |
| 68 | Cnih2 | 13909 | -0.197 | -0.1280 | No |
| 69 | Ybx2 | 14012 | -0.202 | -0.1241 | No |
| 70 | Septin4 | 14034 | -0.203 | -0.1159 | No |
| 71 | Ide | 14170 | -0.208 | -0.1135 | No |
| 72 | Braf | 14936 | -0.247 | -0.1420 | No |
| 73 | Jam3 | 14993 | -0.250 | -0.1335 | No |
| 74 | Arl4a | 15185 | -0.260 | -0.1316 | No |
| 75 | Cdkn3 | 15261 | -0.264 | -0.1235 | No |
| 76 | Tekt2 | 15808 | -0.295 | -0.1385 | No |
| 77 | Pebp1 | 15821 | -0.296 | -0.1256 | No |
| 78 | Grm8 | 16044 | -0.310 | -0.1230 | No |
| 79 | Spata6 | 16243 | -0.323 | -0.1186 | No |
| 80 | Cftr | 16359 | -0.331 | -0.1095 | No |
| 81 | Dmc1 | 16507 | -0.342 | -0.1015 | No |
| 82 | Tsn | 16674 | -0.352 | -0.0941 | No |
| 83 | Ddx4 | 16688 | -0.353 | -0.0787 | No |
| 84 | Gfi1 | 16876 | -0.367 | -0.0717 | No |
| 85 | Adam2 | 16907 | -0.370 | -0.0564 | No |
| 86 | Ddx25 | 17371 | -0.407 | -0.0619 | No |
| 87 | Acrbp | 17603 | -0.427 | -0.0544 | No |
| 88 | Dmrt1 | 17877 | -0.458 | -0.0478 | No |
| 89 | Psmg1 | 17881 | -0.458 | -0.0270 | No |
| 90 | Taldo1 | 18052 | -0.481 | -0.0139 | No |
| 91 | Art3 | 18262 | -0.511 | -0.0015 | No |
| 92 | Gstm5 | 18543 | -0.557 | 0.0093 | No |
| 93 | Il12rb2 | 18925 | -0.666 | 0.0199 | No |