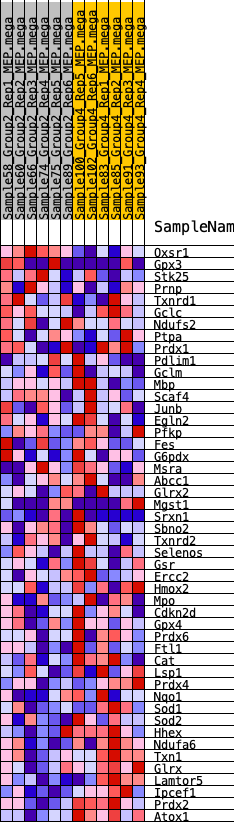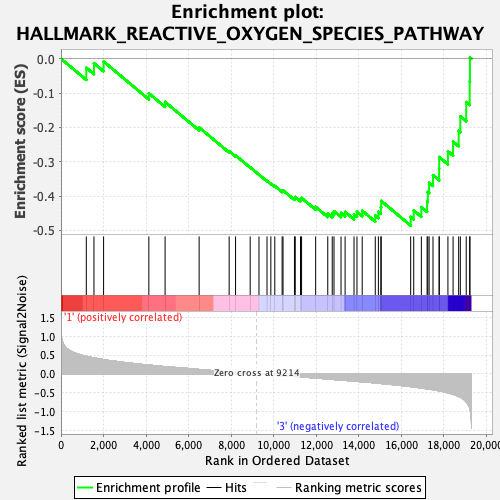
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_REACTIVE_OXYGEN_SPECIES_PATHWAY |
| Enrichment Score (ES) | -0.48598653 |
| Normalized Enrichment Score (NES) | -1.5025195 |
| Nominal p-value | 0.049407113 |
| FDR q-value | 0.969543 |
| FWER p-Value | 0.413 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Oxsr1 | 1190 | 0.469 | -0.0264 | No |
| 2 | Gpx3 | 1550 | 0.422 | -0.0132 | No |
| 3 | Stk25 | 2003 | 0.379 | -0.0081 | No |
| 4 | Prnp | 4132 | 0.235 | -0.1008 | No |
| 5 | Txnrd1 | 4896 | 0.192 | -0.1259 | No |
| 6 | Gclc | 6497 | 0.118 | -0.2001 | No |
| 7 | Ndufs2 | 7909 | 0.055 | -0.2692 | No |
| 8 | Ptpa | 8210 | 0.043 | -0.2815 | No |
| 9 | Prdx1 | 8898 | 0.013 | -0.3162 | No |
| 10 | Pdlim1 | 9312 | -0.000 | -0.3376 | No |
| 11 | Gclm | 9690 | -0.017 | -0.3559 | No |
| 12 | Mbp | 9870 | -0.025 | -0.3633 | No |
| 13 | Scaf4 | 10056 | -0.032 | -0.3705 | No |
| 14 | Junb | 10401 | -0.047 | -0.3849 | No |
| 15 | Egln2 | 10443 | -0.049 | -0.3833 | No |
| 16 | Pfkp | 10981 | -0.069 | -0.4060 | No |
| 17 | Fes | 11018 | -0.071 | -0.4025 | No |
| 18 | G6pdx | 11265 | -0.081 | -0.4092 | No |
| 19 | Msra | 11315 | -0.083 | -0.4055 | No |
| 20 | Abcc1 | 11978 | -0.109 | -0.4316 | No |
| 21 | Glrx2 | 12547 | -0.134 | -0.4510 | No |
| 22 | Mgst1 | 12758 | -0.144 | -0.4511 | No |
| 23 | Srxn1 | 12841 | -0.148 | -0.4442 | No |
| 24 | Sbno2 | 13171 | -0.162 | -0.4490 | No |
| 25 | Txnrd2 | 13363 | -0.171 | -0.4461 | No |
| 26 | Selenos | 13782 | -0.191 | -0.4534 | No |
| 27 | Gsr | 13921 | -0.197 | -0.4457 | No |
| 28 | Ercc2 | 14168 | -0.208 | -0.4428 | No |
| 29 | Hmox2 | 14782 | -0.238 | -0.4566 | No |
| 30 | Mpo | 14928 | -0.247 | -0.4456 | No |
| 31 | Cdkn2d | 15045 | -0.253 | -0.4325 | No |
| 32 | Gpx4 | 15060 | -0.254 | -0.4140 | No |
| 33 | Prdx6 | 16447 | -0.337 | -0.4606 | Yes |
| 34 | Ftl1 | 16590 | -0.347 | -0.4418 | Yes |
| 35 | Cat | 16949 | -0.374 | -0.4321 | Yes |
| 36 | Lsp1 | 17220 | -0.395 | -0.4164 | Yes |
| 37 | Prdx4 | 17252 | -0.397 | -0.3881 | Yes |
| 38 | Nqo1 | 17315 | -0.401 | -0.3611 | Yes |
| 39 | Sod1 | 17497 | -0.417 | -0.3390 | Yes |
| 40 | Sod2 | 17786 | -0.449 | -0.3201 | Yes |
| 41 | Hhex | 17790 | -0.450 | -0.2864 | Yes |
| 42 | Ndufa6 | 18199 | -0.501 | -0.2698 | Yes |
| 43 | Txn1 | 18442 | -0.540 | -0.2416 | Yes |
| 44 | Glrx | 18704 | -0.597 | -0.2102 | Yes |
| 45 | Lamtor5 | 18783 | -0.618 | -0.1676 | Yes |
| 46 | Ipcef1 | 19060 | -0.738 | -0.1263 | Yes |
| 47 | Prdx2 | 19223 | -0.906 | -0.0663 | Yes |
| 48 | Atox1 | 19235 | -0.937 | 0.0037 | Yes |