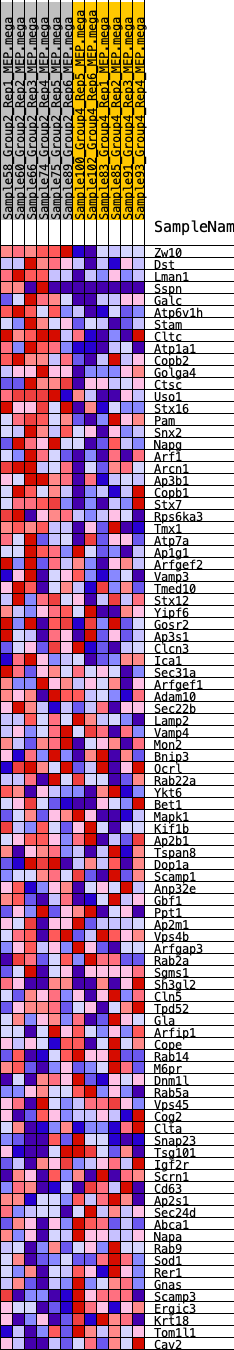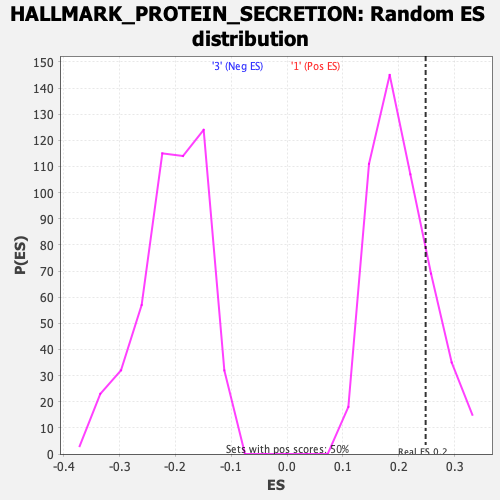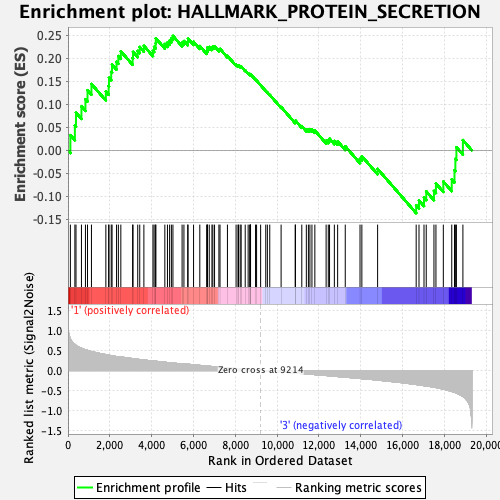
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.24820344 |
| Normalized Enrichment Score (NES) | 1.2259291 |
| Nominal p-value | 0.208 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.85 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zw10 | 114 | 0.794 | 0.0328 | Yes |
| 2 | Dst | 329 | 0.654 | 0.0536 | Yes |
| 3 | Lman1 | 382 | 0.630 | 0.0816 | Yes |
| 4 | Sspn | 643 | 0.559 | 0.0953 | Yes |
| 5 | Galc | 834 | 0.520 | 0.1108 | Yes |
| 6 | Atp6v1h | 933 | 0.506 | 0.1303 | Yes |
| 7 | Stam | 1121 | 0.477 | 0.1438 | Yes |
| 8 | Cltc | 1810 | 0.399 | 0.1275 | Yes |
| 9 | Atp1a1 | 1943 | 0.387 | 0.1395 | Yes |
| 10 | Copb2 | 1960 | 0.384 | 0.1574 | Yes |
| 11 | Golga4 | 2066 | 0.374 | 0.1702 | Yes |
| 12 | Ctsc | 2106 | 0.369 | 0.1861 | Yes |
| 13 | Uso1 | 2321 | 0.350 | 0.1920 | Yes |
| 14 | Stx16 | 2408 | 0.345 | 0.2044 | Yes |
| 15 | Pam | 2526 | 0.338 | 0.2147 | Yes |
| 16 | Snx2 | 3089 | 0.298 | 0.2000 | Yes |
| 17 | Napg | 3109 | 0.297 | 0.2135 | Yes |
| 18 | Arf1 | 3334 | 0.282 | 0.2156 | Yes |
| 19 | Arcn1 | 3430 | 0.275 | 0.2241 | Yes |
| 20 | Ap3b1 | 3628 | 0.263 | 0.2267 | Yes |
| 21 | Copb1 | 4061 | 0.240 | 0.2159 | Yes |
| 22 | Stx7 | 4118 | 0.236 | 0.2245 | Yes |
| 23 | Rps6ka3 | 4191 | 0.232 | 0.2320 | Yes |
| 24 | Tmx1 | 4198 | 0.231 | 0.2430 | Yes |
| 25 | Atp7a | 4630 | 0.208 | 0.2307 | Yes |
| 26 | Ap1g1 | 4755 | 0.200 | 0.2339 | Yes |
| 27 | Arfgef2 | 4858 | 0.194 | 0.2381 | Yes |
| 28 | Vamp3 | 4943 | 0.190 | 0.2429 | Yes |
| 29 | Tmed10 | 5017 | 0.186 | 0.2482 | Yes |
| 30 | Stx12 | 5455 | 0.167 | 0.2336 | No |
| 31 | Yipf6 | 5543 | 0.162 | 0.2370 | No |
| 32 | Gosr2 | 5724 | 0.157 | 0.2353 | No |
| 33 | Ap3s1 | 5739 | 0.156 | 0.2421 | No |
| 34 | Clcn3 | 6003 | 0.142 | 0.2354 | No |
| 35 | Ica1 | 6302 | 0.128 | 0.2261 | No |
| 36 | Sec31a | 6630 | 0.112 | 0.2145 | No |
| 37 | Arfgef1 | 6665 | 0.110 | 0.2181 | No |
| 38 | Adam10 | 6669 | 0.110 | 0.2233 | No |
| 39 | Sec22b | 6754 | 0.106 | 0.2241 | No |
| 40 | Lamp2 | 6882 | 0.100 | 0.2223 | No |
| 41 | Vamp4 | 6926 | 0.097 | 0.2248 | No |
| 42 | Mon2 | 7007 | 0.094 | 0.2253 | No |
| 43 | Bnip3 | 7215 | 0.085 | 0.2186 | No |
| 44 | Ocrl | 7268 | 0.083 | 0.2200 | No |
| 45 | Rab22a | 7623 | 0.067 | 0.2048 | No |
| 46 | Ykt6 | 8036 | 0.050 | 0.1858 | No |
| 47 | Bet1 | 8127 | 0.047 | 0.1834 | No |
| 48 | Mapk1 | 8152 | 0.046 | 0.1844 | No |
| 49 | Kif1b | 8231 | 0.042 | 0.1824 | No |
| 50 | Ap2b1 | 8279 | 0.040 | 0.1819 | No |
| 51 | Tspan8 | 8473 | 0.030 | 0.1733 | No |
| 52 | Dop1a | 8613 | 0.023 | 0.1672 | No |
| 53 | Scamp1 | 8687 | 0.020 | 0.1644 | No |
| 54 | Anp32e | 8720 | 0.019 | 0.1636 | No |
| 55 | Gbf1 | 8722 | 0.019 | 0.1645 | No |
| 56 | Ppt1 | 8728 | 0.019 | 0.1652 | No |
| 57 | Ap2m1 | 8967 | 0.010 | 0.1533 | No |
| 58 | Vps4b | 9016 | 0.008 | 0.1512 | No |
| 59 | Arfgap3 | 9210 | 0.000 | 0.1411 | No |
| 60 | Rab2a | 9455 | -0.007 | 0.1288 | No |
| 61 | Sgms1 | 9532 | -0.010 | 0.1253 | No |
| 62 | Sh3gl2 | 9646 | -0.015 | 0.1202 | No |
| 63 | Cln5 | 10189 | -0.038 | 0.0938 | No |
| 64 | Tpd52 | 10862 | -0.064 | 0.0620 | No |
| 65 | Gla | 10876 | -0.064 | 0.0644 | No |
| 66 | Arfip1 | 11178 | -0.077 | 0.0525 | No |
| 67 | Cope | 11395 | -0.086 | 0.0455 | No |
| 68 | Rab14 | 11494 | -0.089 | 0.0447 | No |
| 69 | M6pr | 11566 | -0.091 | 0.0455 | No |
| 70 | Dnm1l | 11663 | -0.095 | 0.0451 | No |
| 71 | Rab5a | 11802 | -0.102 | 0.0429 | No |
| 72 | Vps45 | 12341 | -0.124 | 0.0209 | No |
| 73 | Cog2 | 12445 | -0.129 | 0.0218 | No |
| 74 | Clta | 12505 | -0.132 | 0.0252 | No |
| 75 | Snap23 | 12735 | -0.143 | 0.0203 | No |
| 76 | Tsg101 | 12892 | -0.150 | 0.0195 | No |
| 77 | Igf2r | 13256 | -0.166 | 0.0087 | No |
| 78 | Scrn1 | 13962 | -0.199 | -0.0183 | No |
| 79 | Cd63 | 14050 | -0.204 | -0.0129 | No |
| 80 | Ap2s1 | 14803 | -0.239 | -0.0403 | No |
| 81 | Sec24d | 16647 | -0.350 | -0.1192 | No |
| 82 | Abca1 | 16782 | -0.361 | -0.1086 | No |
| 83 | Napa | 17021 | -0.379 | -0.1025 | No |
| 84 | Rab9 | 17128 | -0.388 | -0.0891 | No |
| 85 | Sod1 | 17497 | -0.417 | -0.0879 | No |
| 86 | Rer1 | 17592 | -0.427 | -0.0720 | No |
| 87 | Gnas | 17945 | -0.465 | -0.0677 | No |
| 88 | Scamp3 | 18348 | -0.524 | -0.0630 | No |
| 89 | Ergic3 | 18479 | -0.547 | -0.0432 | No |
| 90 | Krt18 | 18521 | -0.553 | -0.0183 | No |
| 91 | Tom1l1 | 18571 | -0.563 | 0.0066 | No |
| 92 | Cav2 | 18881 | -0.649 | 0.0222 | No |