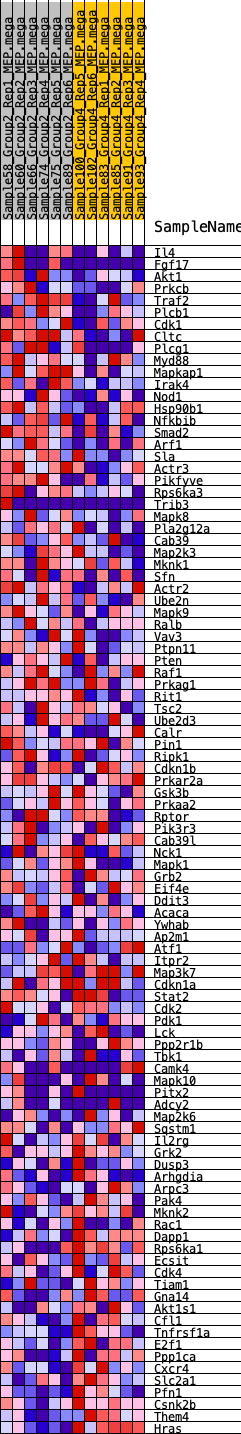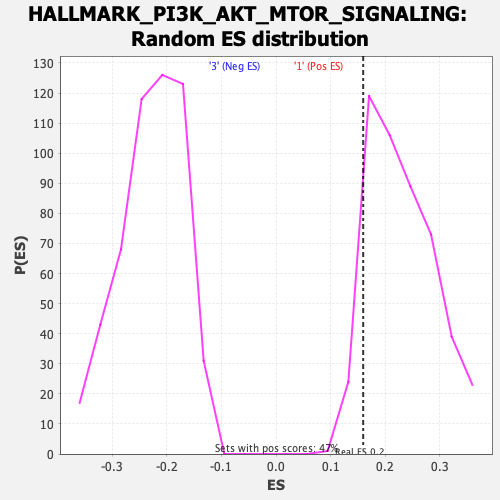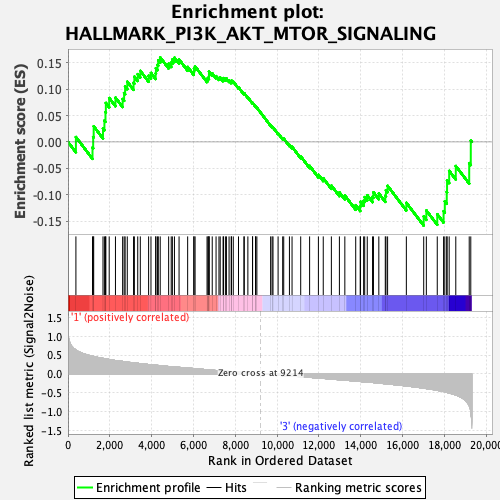
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.15981184 |
| Normalized Enrichment Score (NES) | 0.692964 |
| Nominal p-value | 0.9092827 |
| FDR q-value | 0.9351043 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Il4 | 378 | 0.631 | 0.0093 | Yes |
| 2 | Fgf17 | 1172 | 0.469 | -0.0104 | Yes |
| 3 | Akt1 | 1204 | 0.468 | 0.0094 | Yes |
| 4 | Prkcb | 1228 | 0.464 | 0.0296 | Yes |
| 5 | Traf2 | 1673 | 0.413 | 0.0254 | Yes |
| 6 | Plcb1 | 1741 | 0.406 | 0.0406 | Yes |
| 7 | Cdk1 | 1792 | 0.400 | 0.0563 | Yes |
| 8 | Cltc | 1810 | 0.399 | 0.0738 | Yes |
| 9 | Plcg1 | 1970 | 0.382 | 0.0830 | Yes |
| 10 | Myd88 | 2269 | 0.354 | 0.0838 | Yes |
| 11 | Mapkap1 | 2612 | 0.331 | 0.0812 | Yes |
| 12 | Irak4 | 2683 | 0.328 | 0.0926 | Yes |
| 13 | Nod1 | 2731 | 0.322 | 0.1050 | Yes |
| 14 | Hsp90b1 | 2834 | 0.313 | 0.1140 | Yes |
| 15 | Nfkbib | 3134 | 0.296 | 0.1120 | Yes |
| 16 | Smad2 | 3173 | 0.293 | 0.1235 | Yes |
| 17 | Arf1 | 3334 | 0.282 | 0.1282 | Yes |
| 18 | Sla | 3457 | 0.273 | 0.1344 | Yes |
| 19 | Actr3 | 3854 | 0.249 | 0.1252 | Yes |
| 20 | Pikfyve | 3967 | 0.244 | 0.1305 | Yes |
| 21 | Rps6ka3 | 4191 | 0.232 | 0.1296 | Yes |
| 22 | Trib3 | 4204 | 0.231 | 0.1395 | Yes |
| 23 | Mapk8 | 4282 | 0.227 | 0.1459 | Yes |
| 24 | Pla2g12a | 4317 | 0.224 | 0.1545 | Yes |
| 25 | Cab39 | 4409 | 0.219 | 0.1598 | Yes |
| 26 | Map2k3 | 4812 | 0.196 | 0.1479 | No |
| 27 | Mknk1 | 4935 | 0.190 | 0.1503 | No |
| 28 | Sfn | 4990 | 0.187 | 0.1561 | No |
| 29 | Actr2 | 5089 | 0.183 | 0.1593 | No |
| 30 | Ube2n | 5310 | 0.175 | 0.1559 | No |
| 31 | Mapk9 | 5721 | 0.157 | 0.1418 | No |
| 32 | Ralb | 6012 | 0.142 | 0.1332 | No |
| 33 | Vav3 | 6026 | 0.141 | 0.1390 | No |
| 34 | Ptpn11 | 6076 | 0.138 | 0.1428 | No |
| 35 | Pten | 6648 | 0.111 | 0.1182 | No |
| 36 | Raf1 | 6690 | 0.109 | 0.1210 | No |
| 37 | Prkag1 | 6737 | 0.106 | 0.1235 | No |
| 38 | Rit1 | 6740 | 0.106 | 0.1283 | No |
| 39 | Tsc2 | 6741 | 0.106 | 0.1332 | No |
| 40 | Ube2d3 | 6896 | 0.099 | 0.1297 | No |
| 41 | Calr | 7079 | 0.090 | 0.1244 | No |
| 42 | Pin1 | 7218 | 0.085 | 0.1211 | No |
| 43 | Ripk1 | 7277 | 0.083 | 0.1218 | No |
| 44 | Cdkn1b | 7415 | 0.076 | 0.1182 | No |
| 45 | Prkar2a | 7434 | 0.075 | 0.1207 | No |
| 46 | Gsk3b | 7525 | 0.071 | 0.1193 | No |
| 47 | Prkaa2 | 7575 | 0.069 | 0.1199 | No |
| 48 | Rptor | 7701 | 0.064 | 0.1163 | No |
| 49 | Pik3r3 | 7804 | 0.060 | 0.1138 | No |
| 50 | Cab39l | 7815 | 0.059 | 0.1160 | No |
| 51 | Nck1 | 7902 | 0.056 | 0.1141 | No |
| 52 | Mapk1 | 8152 | 0.046 | 0.1032 | No |
| 53 | Grb2 | 8411 | 0.033 | 0.0913 | No |
| 54 | Eif4e | 8428 | 0.033 | 0.0919 | No |
| 55 | Ddit3 | 8600 | 0.024 | 0.0841 | No |
| 56 | Acaca | 8823 | 0.015 | 0.0733 | No |
| 57 | Ywhab | 8825 | 0.015 | 0.0739 | No |
| 58 | Ap2m1 | 8967 | 0.010 | 0.0671 | No |
| 59 | Atf1 | 8973 | 0.010 | 0.0673 | No |
| 60 | Itpr2 | 9030 | 0.008 | 0.0647 | No |
| 61 | Map3k7 | 9688 | -0.017 | 0.0313 | No |
| 62 | Cdkn1a | 9757 | -0.020 | 0.0287 | No |
| 63 | Stat2 | 9801 | -0.021 | 0.0274 | No |
| 64 | Cdk2 | 10045 | -0.032 | 0.0162 | No |
| 65 | Pdk1 | 10264 | -0.041 | 0.0068 | No |
| 66 | Lck | 10314 | -0.044 | 0.0062 | No |
| 67 | Ppp2r1b | 10587 | -0.054 | -0.0055 | No |
| 68 | Tbk1 | 10712 | -0.059 | -0.0092 | No |
| 69 | Camk4 | 11126 | -0.075 | -0.0273 | No |
| 70 | Mapk10 | 11552 | -0.090 | -0.0453 | No |
| 71 | Pitx2 | 11972 | -0.109 | -0.0620 | No |
| 72 | Adcy2 | 12204 | -0.119 | -0.0686 | No |
| 73 | Map2k6 | 12592 | -0.136 | -0.0825 | No |
| 74 | Sqstm1 | 12976 | -0.154 | -0.0954 | No |
| 75 | Il2rg | 13236 | -0.165 | -0.1013 | No |
| 76 | Grk2 | 13756 | -0.190 | -0.1196 | No |
| 77 | Dusp3 | 13971 | -0.200 | -0.1215 | No |
| 78 | Arhgdia | 13986 | -0.201 | -0.1131 | No |
| 79 | Arpc3 | 14135 | -0.206 | -0.1113 | No |
| 80 | Pak4 | 14186 | -0.209 | -0.1043 | No |
| 81 | Mknk2 | 14308 | -0.214 | -0.1008 | No |
| 82 | Rac1 | 14561 | -0.226 | -0.1035 | No |
| 83 | Dapp1 | 14604 | -0.228 | -0.0952 | No |
| 84 | Rps6ka1 | 14861 | -0.243 | -0.0974 | No |
| 85 | Ecsit | 15170 | -0.259 | -0.1015 | No |
| 86 | Cdk4 | 15202 | -0.261 | -0.0912 | No |
| 87 | Tiam1 | 15281 | -0.265 | -0.0830 | No |
| 88 | Gna14 | 16177 | -0.318 | -0.1150 | No |
| 89 | Akt1s1 | 17007 | -0.378 | -0.1408 | No |
| 90 | Cfl1 | 17133 | -0.388 | -0.1295 | No |
| 91 | Tnfrsf1a | 17655 | -0.433 | -0.1367 | No |
| 92 | E2f1 | 17957 | -0.466 | -0.1310 | No |
| 93 | Ppp1ca | 18016 | -0.475 | -0.1122 | No |
| 94 | Cxcr4 | 18109 | -0.489 | -0.0945 | No |
| 95 | Slc2a1 | 18124 | -0.491 | -0.0727 | No |
| 96 | Pfn1 | 18225 | -0.506 | -0.0547 | No |
| 97 | Csnk2b | 18541 | -0.557 | -0.0455 | No |
| 98 | Them4 | 19183 | -0.845 | -0.0401 | No |
| 99 | Hras | 19258 | -1.011 | 0.0026 | No |