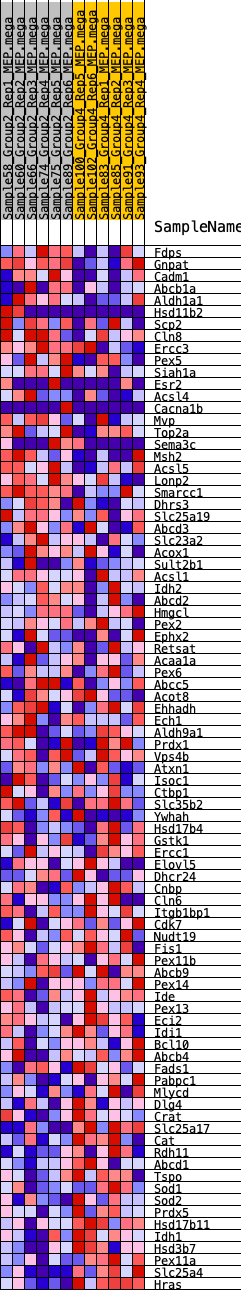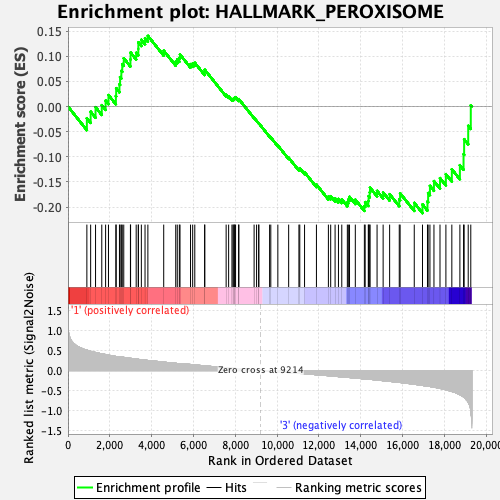
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.21204911 |
| Normalized Enrichment Score (NES) | -0.9533671 |
| Nominal p-value | 0.53831416 |
| FDR q-value | 0.86734843 |
| FWER p-Value | 0.992 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fdps | 902 | 0.511 | -0.0231 | No |
| 2 | Gnpat | 1083 | 0.480 | -0.0101 | No |
| 3 | Cadm1 | 1317 | 0.451 | -0.0012 | No |
| 4 | Abcb1a | 1615 | 0.418 | 0.0028 | No |
| 5 | Aldh1a1 | 1798 | 0.399 | 0.0119 | No |
| 6 | Hsd11b2 | 1935 | 0.387 | 0.0229 | No |
| 7 | Scp2 | 2289 | 0.352 | 0.0209 | No |
| 8 | Cln8 | 2303 | 0.351 | 0.0366 | No |
| 9 | Ercc3 | 2460 | 0.343 | 0.0444 | No |
| 10 | Pex5 | 2490 | 0.341 | 0.0588 | No |
| 11 | Siah1a | 2551 | 0.336 | 0.0713 | No |
| 12 | Esr2 | 2594 | 0.332 | 0.0846 | No |
| 13 | Acsl4 | 2666 | 0.329 | 0.0962 | No |
| 14 | Cacna1b | 2985 | 0.303 | 0.0938 | No |
| 15 | Mvp | 2992 | 0.303 | 0.1076 | No |
| 16 | Top2a | 3260 | 0.287 | 0.1071 | No |
| 17 | Sema3c | 3362 | 0.280 | 0.1148 | No |
| 18 | Msh2 | 3364 | 0.280 | 0.1278 | No |
| 19 | Acsl5 | 3507 | 0.271 | 0.1330 | No |
| 20 | Lonp2 | 3683 | 0.260 | 0.1360 | No |
| 21 | Smarcc1 | 3819 | 0.251 | 0.1407 | No |
| 22 | Dhrs3 | 4576 | 0.211 | 0.1112 | No |
| 23 | Slc25a19 | 5151 | 0.180 | 0.0897 | No |
| 24 | Abcd3 | 5224 | 0.176 | 0.0942 | No |
| 25 | Slc23a2 | 5323 | 0.174 | 0.0971 | No |
| 26 | Acox1 | 5355 | 0.172 | 0.1036 | No |
| 27 | Sult2b1 | 5858 | 0.151 | 0.0845 | No |
| 28 | Acsl1 | 5961 | 0.145 | 0.0859 | No |
| 29 | Idh2 | 6062 | 0.139 | 0.0872 | No |
| 30 | Abcd2 | 6533 | 0.116 | 0.0681 | No |
| 31 | Hmgcl | 6539 | 0.116 | 0.0733 | No |
| 32 | Pex2 | 7561 | 0.070 | 0.0234 | No |
| 33 | Ephx2 | 7680 | 0.065 | 0.0203 | No |
| 34 | Retsat | 7839 | 0.059 | 0.0148 | No |
| 35 | Acaa1a | 7890 | 0.056 | 0.0148 | No |
| 36 | Pex6 | 7930 | 0.055 | 0.0153 | No |
| 37 | Abcc5 | 7948 | 0.054 | 0.0170 | No |
| 38 | Acot8 | 7976 | 0.053 | 0.0180 | No |
| 39 | Ehhadh | 8012 | 0.051 | 0.0186 | No |
| 40 | Ech1 | 8158 | 0.045 | 0.0132 | No |
| 41 | Aldh9a1 | 8169 | 0.045 | 0.0147 | No |
| 42 | Prdx1 | 8898 | 0.013 | -0.0225 | No |
| 43 | Vps4b | 9016 | 0.008 | -0.0282 | No |
| 44 | Atxn1 | 9118 | 0.004 | -0.0333 | No |
| 45 | Isoc1 | 9123 | 0.004 | -0.0333 | No |
| 46 | Ctbp1 | 9639 | -0.015 | -0.0594 | No |
| 47 | Slc35b2 | 9696 | -0.017 | -0.0615 | No |
| 48 | Ywhah | 10033 | -0.031 | -0.0775 | No |
| 49 | Hsd17b4 | 10550 | -0.053 | -0.1019 | No |
| 50 | Gstk1 | 11041 | -0.072 | -0.1241 | No |
| 51 | Ercc1 | 11082 | -0.073 | -0.1227 | No |
| 52 | Elovl5 | 11310 | -0.083 | -0.1307 | No |
| 53 | Dhcr24 | 11878 | -0.105 | -0.1553 | No |
| 54 | Cnbp | 12450 | -0.129 | -0.1790 | No |
| 55 | Cln6 | 12560 | -0.134 | -0.1784 | No |
| 56 | Itgb1bp1 | 12770 | -0.144 | -0.1825 | No |
| 57 | Cdk7 | 12931 | -0.152 | -0.1838 | No |
| 58 | Nudt19 | 13090 | -0.158 | -0.1846 | No |
| 59 | Fis1 | 13356 | -0.171 | -0.1905 | No |
| 60 | Pex11b | 13403 | -0.173 | -0.1848 | No |
| 61 | Abcb9 | 13460 | -0.175 | -0.1796 | No |
| 62 | Pex14 | 13737 | -0.189 | -0.1851 | No |
| 63 | Ide | 14170 | -0.208 | -0.1979 | Yes |
| 64 | Pex13 | 14208 | -0.210 | -0.1901 | Yes |
| 65 | Eci2 | 14358 | -0.217 | -0.1877 | Yes |
| 66 | Idi1 | 14372 | -0.217 | -0.1783 | Yes |
| 67 | Bcl10 | 14425 | -0.220 | -0.1708 | Yes |
| 68 | Abcb4 | 14440 | -0.220 | -0.1613 | Yes |
| 69 | Fads1 | 14777 | -0.238 | -0.1677 | Yes |
| 70 | Pabpc1 | 15067 | -0.255 | -0.1709 | Yes |
| 71 | Mlycd | 15378 | -0.271 | -0.1744 | Yes |
| 72 | Dlg4 | 15838 | -0.297 | -0.1844 | Yes |
| 73 | Crat | 15879 | -0.300 | -0.1726 | Yes |
| 74 | Slc25a17 | 16555 | -0.345 | -0.1916 | Yes |
| 75 | Cat | 16949 | -0.374 | -0.1946 | Yes |
| 76 | Rdh11 | 17187 | -0.392 | -0.1887 | Yes |
| 77 | Abcd1 | 17219 | -0.395 | -0.1720 | Yes |
| 78 | Tspo | 17305 | -0.400 | -0.1577 | Yes |
| 79 | Sod1 | 17497 | -0.417 | -0.1483 | Yes |
| 80 | Sod2 | 17786 | -0.449 | -0.1424 | Yes |
| 81 | Prdx5 | 18068 | -0.483 | -0.1345 | Yes |
| 82 | Hsd17b11 | 18352 | -0.525 | -0.1248 | Yes |
| 83 | Idh1 | 18737 | -0.604 | -0.1166 | Yes |
| 84 | Hsd3b7 | 18912 | -0.661 | -0.0949 | Yes |
| 85 | Pex11a | 18940 | -0.672 | -0.0650 | Yes |
| 86 | Slc25a4 | 19135 | -0.791 | -0.0382 | Yes |
| 87 | Hras | 19258 | -1.011 | 0.0025 | Yes |