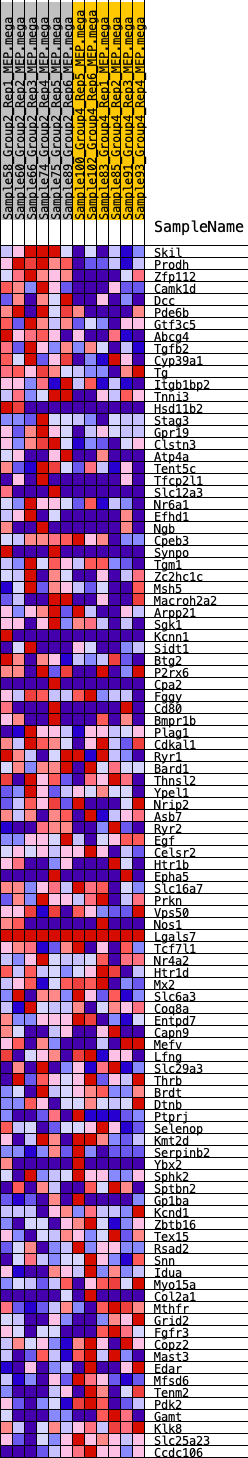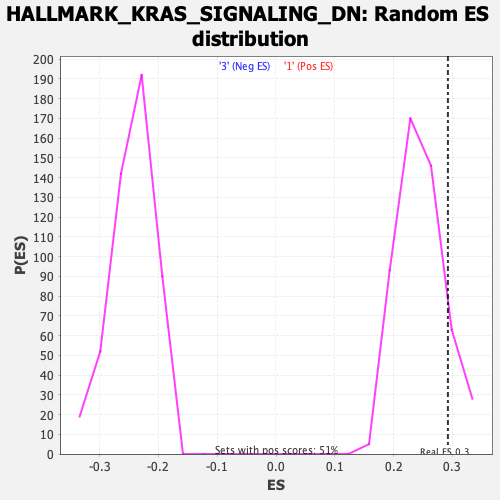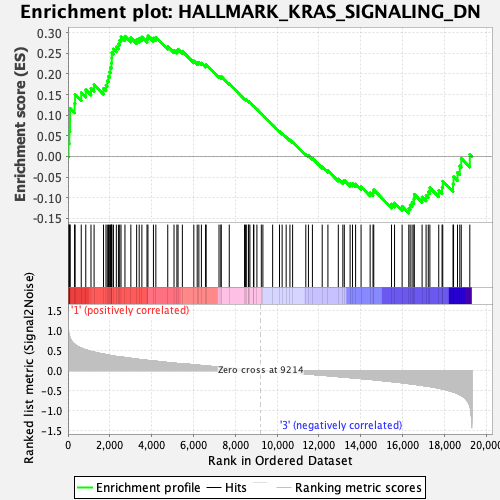
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.2924562 |
| Normalized Enrichment Score (NES) | 1.1895652 |
| Nominal p-value | 0.12871288 |
| FDR q-value | 0.9802313 |
| FWER p-Value | 0.885 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Skil | 27 | 0.964 | 0.0325 | Yes |
| 2 | Prodh | 66 | 0.872 | 0.0611 | Yes |
| 3 | Zfp112 | 94 | 0.817 | 0.0884 | Yes |
| 4 | Camk1d | 96 | 0.815 | 0.1169 | Yes |
| 5 | Dcc | 314 | 0.658 | 0.1287 | Yes |
| 6 | Pde6b | 340 | 0.649 | 0.1502 | Yes |
| 7 | Gtf3c5 | 633 | 0.562 | 0.1547 | Yes |
| 8 | Abcg4 | 850 | 0.518 | 0.1617 | Yes |
| 9 | Tgfb2 | 1099 | 0.478 | 0.1656 | Yes |
| 10 | Cyp39a1 | 1247 | 0.461 | 0.1741 | Yes |
| 11 | Tg | 1698 | 0.410 | 0.1651 | Yes |
| 12 | Itgb1bp2 | 1816 | 0.398 | 0.1730 | Yes |
| 13 | Tnni3 | 1878 | 0.393 | 0.1836 | Yes |
| 14 | Hsd11b2 | 1935 | 0.387 | 0.1943 | Yes |
| 15 | Stag3 | 1991 | 0.380 | 0.2047 | Yes |
| 16 | Gpr19 | 2031 | 0.377 | 0.2159 | Yes |
| 17 | Clstn3 | 2074 | 0.373 | 0.2269 | Yes |
| 18 | Atp4a | 2091 | 0.371 | 0.2390 | Yes |
| 19 | Tent5c | 2095 | 0.370 | 0.2519 | Yes |
| 20 | Tfcp2l1 | 2173 | 0.363 | 0.2606 | Yes |
| 21 | Slc12a3 | 2311 | 0.350 | 0.2658 | Yes |
| 22 | Nr6a1 | 2411 | 0.344 | 0.2727 | Yes |
| 23 | Efhd1 | 2471 | 0.342 | 0.2817 | Yes |
| 24 | Ngb | 2534 | 0.337 | 0.2903 | Yes |
| 25 | Cpeb3 | 2723 | 0.323 | 0.2918 | Yes |
| 26 | Synpo | 3005 | 0.303 | 0.2878 | Yes |
| 27 | Tgm1 | 3282 | 0.285 | 0.2835 | Yes |
| 28 | Zc2hc1c | 3404 | 0.276 | 0.2869 | Yes |
| 29 | Msh5 | 3532 | 0.269 | 0.2897 | Yes |
| 30 | Macroh2a2 | 3778 | 0.254 | 0.2859 | Yes |
| 31 | Arpp21 | 3822 | 0.251 | 0.2925 | Yes |
| 32 | Sgk1 | 4085 | 0.238 | 0.2872 | No |
| 33 | Kcnn1 | 4200 | 0.231 | 0.2893 | No |
| 34 | Sidt1 | 4769 | 0.199 | 0.2668 | No |
| 35 | Btg2 | 5069 | 0.184 | 0.2576 | No |
| 36 | P2rx6 | 5207 | 0.177 | 0.2567 | No |
| 37 | Cpa2 | 5261 | 0.176 | 0.2601 | No |
| 38 | Fggy | 5468 | 0.166 | 0.2552 | No |
| 39 | Cd80 | 6015 | 0.142 | 0.2318 | No |
| 40 | Bmpr1b | 6177 | 0.133 | 0.2281 | No |
| 41 | Plag1 | 6257 | 0.129 | 0.2285 | No |
| 42 | Cdkal1 | 6378 | 0.124 | 0.2266 | No |
| 43 | Ryr1 | 6577 | 0.114 | 0.2203 | No |
| 44 | Bard1 | 6605 | 0.112 | 0.2229 | No |
| 45 | Thnsl2 | 7220 | 0.085 | 0.1939 | No |
| 46 | Ypel1 | 7314 | 0.081 | 0.1919 | No |
| 47 | Nrip2 | 7320 | 0.081 | 0.1944 | No |
| 48 | Asb7 | 7710 | 0.064 | 0.1764 | No |
| 49 | Ryr2 | 8440 | 0.032 | 0.1396 | No |
| 50 | Egf | 8490 | 0.029 | 0.1381 | No |
| 51 | Celsr2 | 8525 | 0.028 | 0.1373 | No |
| 52 | Htr1b | 8624 | 0.023 | 0.1330 | No |
| 53 | Epha5 | 8642 | 0.022 | 0.1328 | No |
| 54 | Slc16a7 | 8697 | 0.020 | 0.1307 | No |
| 55 | Prkn | 8868 | 0.014 | 0.1224 | No |
| 56 | Vps50 | 8887 | 0.014 | 0.1219 | No |
| 57 | Nos1 | 9032 | 0.008 | 0.1147 | No |
| 58 | Lgals7 | 9243 | 0.000 | 0.1038 | No |
| 59 | Tcf7l1 | 9318 | -0.001 | 0.0999 | No |
| 60 | Nr4a2 | 9784 | -0.021 | 0.0765 | No |
| 61 | Htr1d | 10112 | -0.034 | 0.0607 | No |
| 62 | Mx2 | 10238 | -0.040 | 0.0556 | No |
| 63 | Slc6a3 | 10431 | -0.048 | 0.0473 | No |
| 64 | Coq8a | 10611 | -0.055 | 0.0399 | No |
| 65 | Entpd7 | 10739 | -0.060 | 0.0353 | No |
| 66 | Capn9 | 11370 | -0.085 | 0.0055 | No |
| 67 | Mefv | 11496 | -0.090 | 0.0022 | No |
| 68 | Lfng | 11686 | -0.096 | -0.0043 | No |
| 69 | Slc29a3 | 12154 | -0.117 | -0.0245 | No |
| 70 | Thrb | 12427 | -0.128 | -0.0342 | No |
| 71 | Brdt | 12924 | -0.152 | -0.0547 | No |
| 72 | Dtnb | 13141 | -0.161 | -0.0603 | No |
| 73 | Ptprj | 13211 | -0.164 | -0.0581 | No |
| 74 | Selenop | 13488 | -0.177 | -0.0663 | No |
| 75 | Kmt2d | 13602 | -0.182 | -0.0658 | No |
| 76 | Serpinb2 | 13751 | -0.190 | -0.0668 | No |
| 77 | Ybx2 | 14012 | -0.202 | -0.0733 | No |
| 78 | Sphk2 | 14446 | -0.220 | -0.0881 | No |
| 79 | Sptbn2 | 14584 | -0.228 | -0.0872 | No |
| 80 | Gp1ba | 14619 | -0.230 | -0.0809 | No |
| 81 | Kcnd1 | 15468 | -0.276 | -0.1154 | No |
| 82 | Zbtb16 | 15613 | -0.285 | -0.1129 | No |
| 83 | Tex15 | 15975 | -0.305 | -0.1210 | No |
| 84 | Rsad2 | 16294 | -0.326 | -0.1261 | No |
| 85 | Snn | 16358 | -0.330 | -0.1177 | No |
| 86 | Idua | 16451 | -0.337 | -0.1107 | No |
| 87 | Myo15a | 16539 | -0.344 | -0.1031 | No |
| 88 | Col2a1 | 16553 | -0.345 | -0.0917 | No |
| 89 | Mthfr | 16930 | -0.372 | -0.0982 | No |
| 90 | Grid2 | 17121 | -0.387 | -0.0945 | No |
| 91 | Fgfr3 | 17226 | -0.396 | -0.0860 | No |
| 92 | Copz2 | 17302 | -0.400 | -0.0759 | No |
| 93 | Mast3 | 17726 | -0.441 | -0.0824 | No |
| 94 | Edar | 17885 | -0.459 | -0.0745 | No |
| 95 | Mfsd6 | 17918 | -0.463 | -0.0599 | No |
| 96 | Tenm2 | 18409 | -0.534 | -0.0667 | No |
| 97 | Pdk2 | 18434 | -0.539 | -0.0490 | No |
| 98 | Gamt | 18623 | -0.575 | -0.0386 | No |
| 99 | Klk8 | 18732 | -0.604 | -0.0230 | No |
| 100 | Slc25a23 | 18802 | -0.623 | -0.0047 | No |
| 101 | Ccdc106 | 19211 | -0.882 | 0.0050 | No |