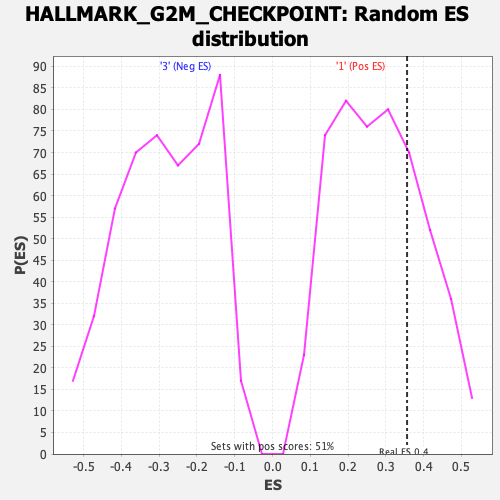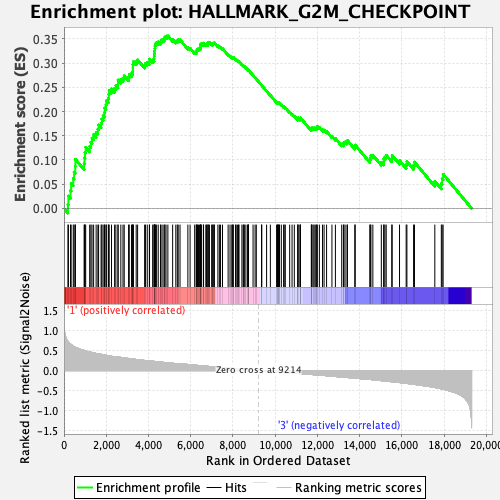
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_G2M_CHECKPOINT |
| Enrichment Score (ES) | 0.35611695 |
| Normalized Enrichment Score (NES) | 1.2668464 |
| Nominal p-value | 0.27667984 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.801 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Nek2 | 187 | 0.727 | 0.0086 | Yes |
| 2 | Srsf1 | 209 | 0.714 | 0.0255 | Yes |
| 3 | Hspa8 | 309 | 0.660 | 0.0370 | Yes |
| 4 | Kif23 | 338 | 0.650 | 0.0520 | Yes |
| 5 | Hnrnpu | 438 | 0.608 | 0.0621 | Yes |
| 6 | Lmnb1 | 478 | 0.598 | 0.0752 | Yes |
| 7 | Upf1 | 533 | 0.583 | 0.0871 | Yes |
| 8 | Xpo1 | 539 | 0.582 | 0.1015 | Yes |
| 9 | Prc1 | 953 | 0.502 | 0.0926 | Yes |
| 10 | Aurka | 972 | 0.498 | 0.1042 | Yes |
| 11 | Plk1 | 994 | 0.494 | 0.1156 | Yes |
| 12 | Sfpq | 1027 | 0.490 | 0.1263 | Yes |
| 13 | Kif2c | 1217 | 0.465 | 0.1281 | Yes |
| 14 | Rad21 | 1273 | 0.457 | 0.1368 | Yes |
| 15 | Cdkn2c | 1327 | 0.450 | 0.1454 | Yes |
| 16 | Amd1 | 1403 | 0.441 | 0.1526 | Yes |
| 17 | Kpnb1 | 1522 | 0.426 | 0.1572 | Yes |
| 18 | Bcl3 | 1605 | 0.420 | 0.1635 | Yes |
| 19 | Hnrnpd | 1638 | 0.416 | 0.1723 | Yes |
| 20 | Ttk | 1757 | 0.405 | 0.1764 | Yes |
| 21 | Cdk1 | 1792 | 0.400 | 0.1847 | Yes |
| 22 | Chaf1a | 1853 | 0.395 | 0.1915 | Yes |
| 23 | Racgap1 | 1920 | 0.389 | 0.1979 | Yes |
| 24 | Hmmr | 1928 | 0.388 | 0.2073 | Yes |
| 25 | Incenp | 1989 | 0.380 | 0.2138 | Yes |
| 26 | Cdc25b | 2006 | 0.379 | 0.2225 | Yes |
| 27 | Bub1 | 2098 | 0.370 | 0.2271 | Yes |
| 28 | Kif11 | 2119 | 0.368 | 0.2353 | Yes |
| 29 | Numa1 | 2137 | 0.367 | 0.2437 | Yes |
| 30 | Ccna2 | 2251 | 0.355 | 0.2468 | Yes |
| 31 | Tpx2 | 2397 | 0.346 | 0.2479 | Yes |
| 32 | Dmd | 2457 | 0.343 | 0.2535 | Yes |
| 33 | Espl1 | 2558 | 0.335 | 0.2567 | Yes |
| 34 | Tent4a | 2560 | 0.335 | 0.2651 | Yes |
| 35 | Cenpf | 2692 | 0.327 | 0.2665 | Yes |
| 36 | Ccnf | 2794 | 0.317 | 0.2693 | Yes |
| 37 | Cenpe | 2852 | 0.312 | 0.2741 | Yes |
| 38 | Map3k20 | 3059 | 0.301 | 0.2710 | Yes |
| 39 | Cul3 | 3090 | 0.298 | 0.2769 | Yes |
| 40 | Dr1 | 3196 | 0.291 | 0.2788 | Yes |
| 41 | Tacc3 | 3255 | 0.287 | 0.2830 | Yes |
| 42 | Top2a | 3260 | 0.287 | 0.2900 | Yes |
| 43 | Prpf4b | 3261 | 0.287 | 0.2973 | Yes |
| 44 | Efna5 | 3287 | 0.285 | 0.3031 | Yes |
| 45 | Cdc27 | 3419 | 0.276 | 0.3033 | Yes |
| 46 | Mcm6 | 3486 | 0.271 | 0.3067 | Yes |
| 47 | Smarcc1 | 3819 | 0.251 | 0.2956 | Yes |
| 48 | Uck2 | 3863 | 0.248 | 0.2996 | Yes |
| 49 | Slc12a2 | 3944 | 0.245 | 0.3016 | Yes |
| 50 | Meis2 | 4042 | 0.241 | 0.3026 | Yes |
| 51 | Cul5 | 4051 | 0.240 | 0.3083 | Yes |
| 52 | Kif4 | 4199 | 0.231 | 0.3064 | Yes |
| 53 | Lbr | 4267 | 0.228 | 0.3087 | Yes |
| 54 | Smad3 | 4271 | 0.227 | 0.3143 | Yes |
| 55 | Kif22 | 4278 | 0.227 | 0.3197 | Yes |
| 56 | Kif15 | 4290 | 0.226 | 0.3248 | Yes |
| 57 | Cul4a | 4300 | 0.226 | 0.3300 | Yes |
| 58 | Aurkb | 4310 | 0.225 | 0.3353 | Yes |
| 59 | Dbf4 | 4328 | 0.223 | 0.3400 | Yes |
| 60 | Fbxo5 | 4386 | 0.220 | 0.3426 | Yes |
| 61 | Ilf3 | 4463 | 0.216 | 0.3441 | Yes |
| 62 | Slc7a5 | 4571 | 0.212 | 0.3438 | Yes |
| 63 | Chmp1a | 4598 | 0.210 | 0.3477 | Yes |
| 64 | Sap30 | 4686 | 0.204 | 0.3483 | Yes |
| 65 | Mtf2 | 4748 | 0.200 | 0.3502 | Yes |
| 66 | Ube2c | 4777 | 0.198 | 0.3537 | Yes |
| 67 | Syncrip | 4832 | 0.195 | 0.3558 | Yes |
| 68 | Suv39h1 | 4920 | 0.191 | 0.3561 | Yes |
| 69 | Tfdp1 | 5146 | 0.180 | 0.3489 | No |
| 70 | Smc1a | 5296 | 0.176 | 0.3455 | No |
| 71 | Rad54l | 5367 | 0.172 | 0.3462 | No |
| 72 | Tmpo | 5408 | 0.169 | 0.3484 | No |
| 73 | G3bp1 | 5495 | 0.165 | 0.3480 | No |
| 74 | Knl1 | 5867 | 0.150 | 0.3324 | No |
| 75 | Mki67 | 5972 | 0.144 | 0.3306 | No |
| 76 | Odf2 | 6190 | 0.133 | 0.3226 | No |
| 77 | Kif5b | 6261 | 0.129 | 0.3222 | No |
| 78 | Birc5 | 6280 | 0.129 | 0.3245 | No |
| 79 | Nsd2 | 6297 | 0.128 | 0.3269 | No |
| 80 | Cdc25a | 6321 | 0.127 | 0.3289 | No |
| 81 | Pafah1b1 | 6369 | 0.124 | 0.3296 | No |
| 82 | Ewsr1 | 6421 | 0.122 | 0.3300 | No |
| 83 | Pole | 6461 | 0.120 | 0.3310 | No |
| 84 | Hira | 6467 | 0.119 | 0.3337 | No |
| 85 | Chek1 | 6469 | 0.119 | 0.3367 | No |
| 86 | Cul1 | 6477 | 0.119 | 0.3393 | No |
| 87 | Plk4 | 6517 | 0.117 | 0.3402 | No |
| 88 | Bard1 | 6605 | 0.112 | 0.3385 | No |
| 89 | Hif1a | 6610 | 0.112 | 0.3412 | No |
| 90 | Ddx39a | 6719 | 0.107 | 0.3382 | No |
| 91 | Prim2 | 6746 | 0.106 | 0.3395 | No |
| 92 | Cenpa | 6815 | 0.103 | 0.3386 | No |
| 93 | Mcm2 | 6820 | 0.103 | 0.3410 | No |
| 94 | Wrn | 6830 | 0.102 | 0.3431 | No |
| 95 | H2bc12 | 6889 | 0.099 | 0.3425 | No |
| 96 | Tra2b | 6993 | 0.094 | 0.3395 | No |
| 97 | Nup98 | 7046 | 0.092 | 0.3391 | No |
| 98 | Mapk14 | 7053 | 0.091 | 0.3411 | No |
| 99 | Nasp | 7097 | 0.090 | 0.3411 | No |
| 100 | Polq | 7124 | 0.089 | 0.3420 | No |
| 101 | Odc1 | 7289 | 0.082 | 0.3355 | No |
| 102 | Kif20b | 7360 | 0.079 | 0.3338 | No |
| 103 | Cdkn1b | 7415 | 0.076 | 0.3329 | No |
| 104 | Tle3 | 7508 | 0.071 | 0.3299 | No |
| 105 | Ccnd1 | 7774 | 0.061 | 0.3176 | No |
| 106 | Abl1 | 7864 | 0.057 | 0.3144 | No |
| 107 | Cdc7 | 7947 | 0.054 | 0.3114 | No |
| 108 | Arid4a | 7989 | 0.052 | 0.3106 | No |
| 109 | Rad23b | 8007 | 0.052 | 0.3110 | No |
| 110 | Ccnt1 | 8014 | 0.051 | 0.3120 | No |
| 111 | Rps6ka5 | 8125 | 0.047 | 0.3074 | No |
| 112 | Pds5b | 8154 | 0.045 | 0.3071 | No |
| 113 | Traip | 8214 | 0.043 | 0.3051 | No |
| 114 | Smc2 | 8268 | 0.040 | 0.3034 | No |
| 115 | Ythdc1 | 8290 | 0.039 | 0.3033 | No |
| 116 | Rasal2 | 8427 | 0.033 | 0.2970 | No |
| 117 | Egf | 8490 | 0.029 | 0.2945 | No |
| 118 | Tnpo2 | 8516 | 0.028 | 0.2939 | No |
| 119 | E2f2 | 8541 | 0.027 | 0.2933 | No |
| 120 | Mybl2 | 8615 | 0.023 | 0.2901 | No |
| 121 | Orc5 | 8696 | 0.020 | 0.2864 | No |
| 122 | Ndc80 | 8724 | 0.019 | 0.2854 | No |
| 123 | Ccnb2 | 8729 | 0.019 | 0.2857 | No |
| 124 | Ncl | 8739 | 0.018 | 0.2857 | No |
| 125 | Dkc1 | 8955 | 0.011 | 0.2747 | No |
| 126 | Slc38a1 | 9051 | 0.007 | 0.2699 | No |
| 127 | Atf5 | 9109 | 0.005 | 0.2671 | No |
| 128 | Mcm3 | 9355 | -0.002 | 0.2543 | No |
| 129 | E2f3 | 9369 | -0.003 | 0.2537 | No |
| 130 | Pbk | 9596 | -0.013 | 0.2422 | No |
| 131 | Smc4 | 9774 | -0.020 | 0.2335 | No |
| 132 | Srsf10 | 10069 | -0.032 | 0.2189 | No |
| 133 | Cdc20 | 10115 | -0.034 | 0.2174 | No |
| 134 | Sqle | 10126 | -0.035 | 0.2178 | No |
| 135 | Nusap1 | 10145 | -0.035 | 0.2177 | No |
| 136 | Mnat1 | 10153 | -0.036 | 0.2183 | No |
| 137 | Notch2 | 10184 | -0.038 | 0.2177 | No |
| 138 | Hus1 | 10208 | -0.039 | 0.2174 | No |
| 139 | Prmt5 | 10293 | -0.043 | 0.2141 | No |
| 140 | Ube2s | 10403 | -0.047 | 0.2096 | No |
| 141 | Ss18 | 10429 | -0.048 | 0.2095 | No |
| 142 | Ezh2 | 10482 | -0.050 | 0.2080 | No |
| 143 | Stil | 10689 | -0.058 | 0.1987 | No |
| 144 | Mcm5 | 10802 | -0.061 | 0.1944 | No |
| 145 | Brca2 | 10909 | -0.066 | 0.1905 | No |
| 146 | Jpt1 | 11075 | -0.073 | 0.1838 | No |
| 147 | Stmn1 | 11077 | -0.073 | 0.1855 | No |
| 148 | Snrpd1 | 11085 | -0.073 | 0.1870 | No |
| 149 | Meis1 | 11182 | -0.077 | 0.1840 | No |
| 150 | Stag1 | 11183 | -0.077 | 0.1859 | No |
| 151 | Cks1b | 11202 | -0.078 | 0.1869 | No |
| 152 | Fancc | 11713 | -0.098 | 0.1627 | No |
| 153 | Atrx | 11733 | -0.098 | 0.1642 | No |
| 154 | Lig3 | 11737 | -0.099 | 0.1665 | No |
| 155 | Katna1 | 11798 | -0.101 | 0.1660 | No |
| 156 | Cbx1 | 11847 | -0.104 | 0.1661 | No |
| 157 | Hmgb3 | 11902 | -0.106 | 0.1659 | No |
| 158 | Exo1 | 11970 | -0.109 | 0.1652 | No |
| 159 | Cks2 | 11980 | -0.109 | 0.1675 | No |
| 160 | Gins2 | 11993 | -0.110 | 0.1696 | No |
| 161 | Cdc6 | 12095 | -0.115 | 0.1672 | No |
| 162 | Foxn3 | 12245 | -0.120 | 0.1625 | No |
| 163 | Casp8ap2 | 12319 | -0.123 | 0.1617 | No |
| 164 | Srsf2 | 12437 | -0.128 | 0.1589 | No |
| 165 | Nolc1 | 12691 | -0.140 | 0.1492 | No |
| 166 | Gspt1 | 12855 | -0.148 | 0.1444 | No |
| 167 | Rpa2 | 13154 | -0.161 | 0.1329 | No |
| 168 | Troap | 13234 | -0.165 | 0.1329 | No |
| 169 | E2f4 | 13254 | -0.166 | 0.1361 | No |
| 170 | Top1 | 13313 | -0.169 | 0.1373 | No |
| 171 | Rbl1 | 13404 | -0.173 | 0.1370 | No |
| 172 | Ctcf | 13426 | -0.174 | 0.1403 | No |
| 173 | Pml | 13773 | -0.191 | 0.1270 | No |
| 174 | Orc6 | 13793 | -0.191 | 0.1308 | No |
| 175 | Slc7a1 | 14486 | -0.222 | 0.1002 | No |
| 176 | H2az1 | 14508 | -0.223 | 0.1048 | No |
| 177 | Pttg1 | 14537 | -0.225 | 0.1090 | No |
| 178 | Cdc45 | 14629 | -0.230 | 0.1100 | No |
| 179 | Hmga1b | 15030 | -0.253 | 0.0955 | No |
| 180 | Myc | 15146 | -0.258 | 0.0960 | No |
| 181 | Mad2l1 | 15148 | -0.258 | 0.1024 | No |
| 182 | Cdk4 | 15202 | -0.261 | 0.1062 | No |
| 183 | Cdkn3 | 15261 | -0.264 | 0.1099 | No |
| 184 | Bub3 | 15526 | -0.280 | 0.1031 | No |
| 185 | Nup50 | 15542 | -0.281 | 0.1094 | No |
| 186 | H2ax | 15892 | -0.301 | 0.0988 | No |
| 187 | Marcks | 16206 | -0.320 | 0.0905 | No |
| 188 | Tgfb1 | 16242 | -0.323 | 0.0968 | No |
| 189 | Kmt5a | 16558 | -0.345 | 0.0891 | No |
| 190 | Pola2 | 16600 | -0.348 | 0.0957 | No |
| 191 | Pura | 17564 | -0.425 | 0.0560 | No |
| 192 | Dtymk | 17872 | -0.457 | 0.0515 | No |
| 193 | H2az2 | 17910 | -0.462 | 0.0613 | No |
| 194 | E2f1 | 17957 | -0.466 | 0.0706 | No |