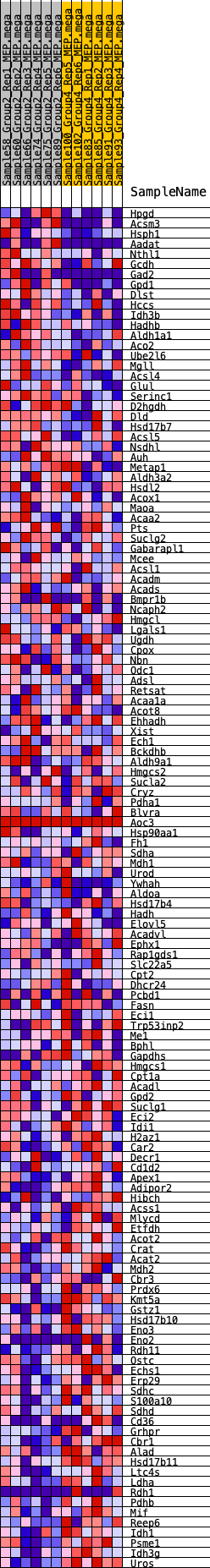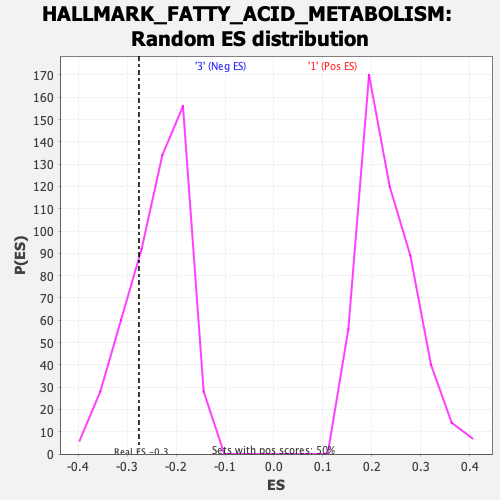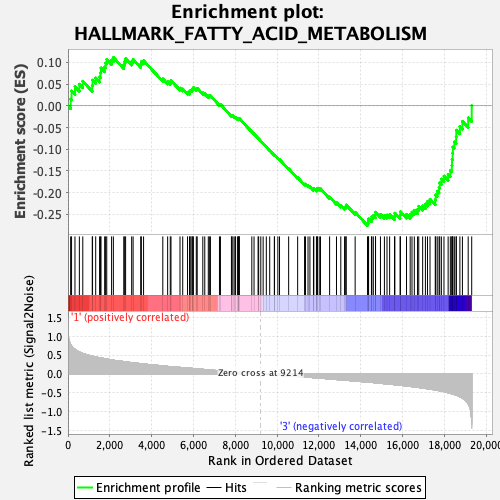
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.27575043 |
| Normalized Enrichment Score (NES) | -1.1629405 |
| Nominal p-value | 0.23809524 |
| FDR q-value | 0.6105484 |
| FWER p-Value | 0.917 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hpgd | 127 | 0.779 | 0.0155 | No |
| 2 | Acsm3 | 170 | 0.738 | 0.0342 | No |
| 3 | Hsph1 | 333 | 0.653 | 0.0443 | No |
| 4 | Aadat | 543 | 0.582 | 0.0498 | No |
| 5 | Nthl1 | 707 | 0.543 | 0.0567 | No |
| 6 | Gcdh | 1160 | 0.471 | 0.0465 | No |
| 7 | Gad2 | 1174 | 0.469 | 0.0591 | No |
| 8 | Gpd1 | 1324 | 0.450 | 0.0641 | No |
| 9 | Dlst | 1510 | 0.428 | 0.0666 | No |
| 10 | Hccs | 1557 | 0.422 | 0.0762 | No |
| 11 | Idh3b | 1576 | 0.421 | 0.0872 | No |
| 12 | Hadhb | 1753 | 0.405 | 0.0895 | No |
| 13 | Aldh1a1 | 1798 | 0.399 | 0.0985 | No |
| 14 | Aco2 | 1852 | 0.395 | 0.1069 | No |
| 15 | Ube2l6 | 2084 | 0.372 | 0.1054 | No |
| 16 | Mgll | 2169 | 0.363 | 0.1113 | No |
| 17 | Acsl4 | 2666 | 0.329 | 0.0948 | No |
| 18 | Glul | 2709 | 0.325 | 0.1018 | No |
| 19 | Serinc1 | 2754 | 0.321 | 0.1086 | No |
| 20 | D2hgdh | 3044 | 0.302 | 0.1021 | No |
| 21 | Dld | 3113 | 0.297 | 0.1070 | No |
| 22 | Hsd17b7 | 3477 | 0.272 | 0.0957 | No |
| 23 | Acsl5 | 3507 | 0.271 | 0.1019 | No |
| 24 | Nsdhl | 3613 | 0.263 | 0.1039 | No |
| 25 | Auh | 4533 | 0.213 | 0.0620 | No |
| 26 | Metap1 | 4764 | 0.199 | 0.0557 | No |
| 27 | Aldh3a2 | 4889 | 0.193 | 0.0546 | No |
| 28 | Hsdl2 | 4924 | 0.191 | 0.0583 | No |
| 29 | Acox1 | 5355 | 0.172 | 0.0407 | No |
| 30 | Maoa | 5485 | 0.165 | 0.0387 | No |
| 31 | Acaa2 | 5715 | 0.157 | 0.0312 | No |
| 32 | Pts | 5811 | 0.153 | 0.0306 | No |
| 33 | Suclg2 | 5825 | 0.152 | 0.0342 | No |
| 34 | Gabarapl1 | 5875 | 0.150 | 0.0359 | No |
| 35 | Mcee | 5932 | 0.146 | 0.0371 | No |
| 36 | Acsl1 | 5961 | 0.145 | 0.0398 | No |
| 37 | Acadm | 5996 | 0.143 | 0.0421 | No |
| 38 | Acads | 6134 | 0.135 | 0.0387 | No |
| 39 | Bmpr1b | 6177 | 0.133 | 0.0403 | No |
| 40 | Ncaph2 | 6445 | 0.121 | 0.0298 | No |
| 41 | Hmgcl | 6539 | 0.116 | 0.0283 | No |
| 42 | Lgals1 | 6708 | 0.108 | 0.0226 | No |
| 43 | Ugdh | 6758 | 0.106 | 0.0230 | No |
| 44 | Cpox | 6810 | 0.103 | 0.0233 | No |
| 45 | Nbn | 7242 | 0.083 | 0.0031 | No |
| 46 | Odc1 | 7289 | 0.082 | 0.0031 | No |
| 47 | Adsl | 7814 | 0.059 | -0.0226 | No |
| 48 | Retsat | 7839 | 0.059 | -0.0222 | No |
| 49 | Acaa1a | 7890 | 0.056 | -0.0232 | No |
| 50 | Acot8 | 7976 | 0.053 | -0.0261 | No |
| 51 | Ehhadh | 8012 | 0.051 | -0.0265 | No |
| 52 | Xist | 8108 | 0.048 | -0.0301 | No |
| 53 | Ech1 | 8158 | 0.045 | -0.0314 | No |
| 54 | Bckdhb | 8166 | 0.045 | -0.0304 | No |
| 55 | Aldh9a1 | 8169 | 0.045 | -0.0293 | No |
| 56 | Hmgcs2 | 8191 | 0.044 | -0.0291 | No |
| 57 | Sucla2 | 8789 | 0.016 | -0.0598 | No |
| 58 | Cryz | 8895 | 0.013 | -0.0649 | No |
| 59 | Pdha1 | 9088 | 0.006 | -0.0747 | No |
| 60 | Blvra | 9116 | 0.004 | -0.0760 | No |
| 61 | Aoc3 | 9218 | 0.000 | -0.0813 | No |
| 62 | Hsp90aa1 | 9330 | -0.001 | -0.0870 | No |
| 63 | Fh1 | 9482 | -0.008 | -0.0947 | No |
| 64 | Sdha | 9644 | -0.015 | -0.1027 | No |
| 65 | Mdh1 | 9865 | -0.024 | -0.1134 | No |
| 66 | Urod | 9874 | -0.025 | -0.1132 | No |
| 67 | Ywhah | 10033 | -0.031 | -0.1205 | No |
| 68 | Aldoa | 10113 | -0.034 | -0.1237 | No |
| 69 | Hsd17b4 | 10550 | -0.053 | -0.1449 | No |
| 70 | Hadh | 10980 | -0.069 | -0.1653 | No |
| 71 | Elovl5 | 11310 | -0.083 | -0.1801 | No |
| 72 | Acadvl | 11356 | -0.084 | -0.1801 | No |
| 73 | Ephx1 | 11476 | -0.089 | -0.1838 | No |
| 74 | Rap1gds1 | 11567 | -0.091 | -0.1859 | No |
| 75 | Slc22a5 | 11736 | -0.099 | -0.1919 | No |
| 76 | Cpt2 | 11753 | -0.099 | -0.1899 | No |
| 77 | Dhcr24 | 11878 | -0.105 | -0.1934 | No |
| 78 | Pcbd1 | 11901 | -0.106 | -0.1915 | No |
| 79 | Fasn | 11926 | -0.108 | -0.1897 | No |
| 80 | Eci1 | 12012 | -0.111 | -0.1910 | No |
| 81 | Trp53inp2 | 12072 | -0.113 | -0.1909 | No |
| 82 | Me1 | 12508 | -0.132 | -0.2098 | No |
| 83 | Bphl | 12837 | -0.148 | -0.2227 | No |
| 84 | Gapdhs | 13043 | -0.156 | -0.2290 | No |
| 85 | Hmgcs1 | 13223 | -0.164 | -0.2337 | No |
| 86 | Cpt1a | 13278 | -0.167 | -0.2317 | No |
| 87 | Acadl | 13302 | -0.168 | -0.2281 | No |
| 88 | Gpd2 | 13729 | -0.188 | -0.2450 | No |
| 89 | Suclg1 | 14319 | -0.214 | -0.2697 | Yes |
| 90 | Eci2 | 14358 | -0.217 | -0.2655 | Yes |
| 91 | Idi1 | 14372 | -0.217 | -0.2600 | Yes |
| 92 | H2az1 | 14508 | -0.223 | -0.2607 | Yes |
| 93 | Car2 | 14527 | -0.224 | -0.2553 | Yes |
| 94 | Decr1 | 14598 | -0.228 | -0.2525 | Yes |
| 95 | Cd1d2 | 14702 | -0.234 | -0.2512 | Yes |
| 96 | Apex1 | 14705 | -0.234 | -0.2447 | Yes |
| 97 | Adipor2 | 14939 | -0.247 | -0.2499 | Yes |
| 98 | Hibch | 15128 | -0.257 | -0.2524 | Yes |
| 99 | Acss1 | 15252 | -0.263 | -0.2513 | Yes |
| 100 | Mlycd | 15378 | -0.271 | -0.2502 | Yes |
| 101 | Etfdh | 15610 | -0.285 | -0.2542 | Yes |
| 102 | Acot2 | 15628 | -0.285 | -0.2469 | Yes |
| 103 | Crat | 15879 | -0.300 | -0.2515 | Yes |
| 104 | Acat2 | 15893 | -0.301 | -0.2437 | Yes |
| 105 | Mdh2 | 16185 | -0.319 | -0.2498 | Yes |
| 106 | Cbr3 | 16360 | -0.331 | -0.2495 | Yes |
| 107 | Prdx6 | 16447 | -0.337 | -0.2444 | Yes |
| 108 | Kmt5a | 16558 | -0.345 | -0.2404 | Yes |
| 109 | Gstz1 | 16713 | -0.355 | -0.2384 | Yes |
| 110 | Hsd17b10 | 16770 | -0.359 | -0.2311 | Yes |
| 111 | Eno3 | 16957 | -0.374 | -0.2302 | Yes |
| 112 | Eno2 | 17088 | -0.385 | -0.2261 | Yes |
| 113 | Rdh11 | 17187 | -0.392 | -0.2201 | Yes |
| 114 | Ostc | 17308 | -0.400 | -0.2150 | Yes |
| 115 | Echs1 | 17559 | -0.424 | -0.2160 | Yes |
| 116 | Erp29 | 17580 | -0.426 | -0.2050 | Yes |
| 117 | Sdhc | 17654 | -0.433 | -0.1965 | Yes |
| 118 | S100a10 | 17741 | -0.443 | -0.1884 | Yes |
| 119 | Sdhd | 17773 | -0.447 | -0.1773 | Yes |
| 120 | Cd36 | 17852 | -0.455 | -0.1685 | Yes |
| 121 | Grhpr | 17976 | -0.469 | -0.1616 | Yes |
| 122 | Cbr1 | 18175 | -0.498 | -0.1578 | Yes |
| 123 | Alad | 18280 | -0.515 | -0.1487 | Yes |
| 124 | Hsd17b11 | 18352 | -0.525 | -0.1375 | Yes |
| 125 | Ltc4s | 18363 | -0.527 | -0.1231 | Yes |
| 126 | Ldha | 18390 | -0.532 | -0.1093 | Yes |
| 127 | Rdh1 | 18400 | -0.533 | -0.0947 | Yes |
| 128 | Pdhb | 18472 | -0.545 | -0.0829 | Yes |
| 129 | Mif | 18558 | -0.560 | -0.0715 | Yes |
| 130 | Reep6 | 18566 | -0.562 | -0.0559 | Yes |
| 131 | Idh1 | 18737 | -0.604 | -0.0477 | Yes |
| 132 | Psme1 | 18858 | -0.642 | -0.0357 | Yes |
| 133 | Idh3g | 19134 | -0.791 | -0.0277 | Yes |
| 134 | Uros | 19302 | -1.292 | 0.0003 | Yes |