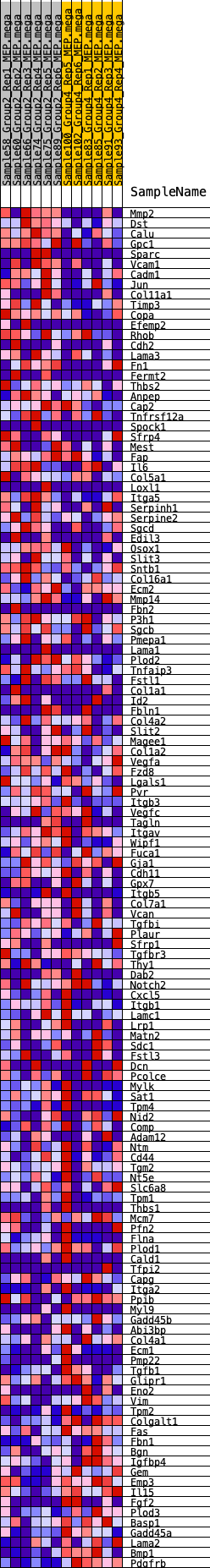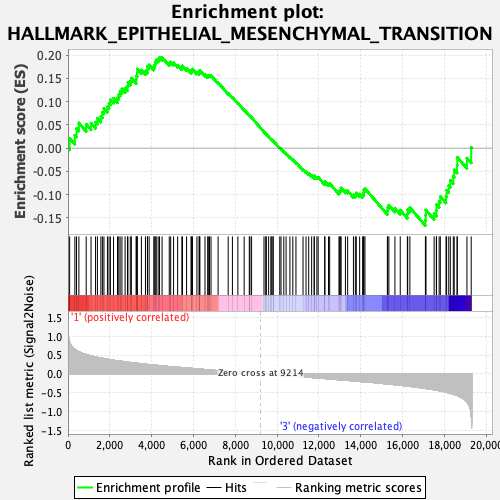
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_EPITHELIAL_MESENCHYMAL_TRANSITION |
| Enrichment Score (ES) | 0.19494964 |
| Normalized Enrichment Score (NES) | 0.7334351 |
| Nominal p-value | 0.8222656 |
| FDR q-value | 0.9338477 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Mmp2 | 78 | 0.847 | 0.0213 | Yes |
| 2 | Dst | 329 | 0.654 | 0.0278 | Yes |
| 3 | Calu | 407 | 0.618 | 0.0423 | Yes |
| 4 | Gpc1 | 520 | 0.588 | 0.0540 | Yes |
| 5 | Sparc | 870 | 0.515 | 0.0512 | Yes |
| 6 | Vcam1 | 1102 | 0.478 | 0.0535 | Yes |
| 7 | Cadm1 | 1317 | 0.451 | 0.0558 | Yes |
| 8 | Jun | 1406 | 0.441 | 0.0645 | Yes |
| 9 | Col11a1 | 1572 | 0.421 | 0.0685 | Yes |
| 10 | Timp3 | 1645 | 0.415 | 0.0771 | Yes |
| 11 | Copa | 1718 | 0.408 | 0.0856 | Yes |
| 12 | Efemp2 | 1885 | 0.392 | 0.0887 | Yes |
| 13 | Rhob | 1961 | 0.384 | 0.0963 | Yes |
| 14 | Cdh2 | 2028 | 0.377 | 0.1041 | Yes |
| 15 | Lama3 | 2177 | 0.362 | 0.1072 | Yes |
| 16 | Fn1 | 2366 | 0.347 | 0.1078 | Yes |
| 17 | Fermt2 | 2427 | 0.343 | 0.1150 | Yes |
| 18 | Thbs2 | 2491 | 0.341 | 0.1219 | Yes |
| 19 | Anpep | 2579 | 0.334 | 0.1273 | Yes |
| 20 | Cap2 | 2730 | 0.322 | 0.1292 | Yes |
| 21 | Tnfrsf12a | 2844 | 0.312 | 0.1326 | Yes |
| 22 | Spock1 | 2865 | 0.311 | 0.1409 | Yes |
| 23 | Sfrp4 | 2978 | 0.304 | 0.1441 | Yes |
| 24 | Mest | 3030 | 0.303 | 0.1505 | Yes |
| 25 | Fap | 3262 | 0.287 | 0.1471 | Yes |
| 26 | Il6 | 3265 | 0.286 | 0.1555 | Yes |
| 27 | Col5a1 | 3308 | 0.283 | 0.1618 | Yes |
| 28 | Loxl1 | 3310 | 0.283 | 0.1703 | Yes |
| 29 | Itga5 | 3509 | 0.271 | 0.1680 | Yes |
| 30 | Serpinh1 | 3703 | 0.258 | 0.1657 | Yes |
| 31 | Serpine2 | 3797 | 0.252 | 0.1684 | Yes |
| 32 | Sgcd | 3801 | 0.252 | 0.1758 | Yes |
| 33 | Edil3 | 3880 | 0.247 | 0.1791 | Yes |
| 34 | Qsox1 | 4102 | 0.237 | 0.1747 | Yes |
| 35 | Slit3 | 4152 | 0.234 | 0.1791 | Yes |
| 36 | Sntb1 | 4166 | 0.233 | 0.1854 | Yes |
| 37 | Col16a1 | 4219 | 0.230 | 0.1896 | Yes |
| 38 | Ecm2 | 4309 | 0.225 | 0.1917 | Yes |
| 39 | Mmp14 | 4376 | 0.221 | 0.1949 | Yes |
| 40 | Fbn2 | 4498 | 0.213 | 0.1949 | Yes |
| 41 | P3h1 | 4841 | 0.195 | 0.1830 | No |
| 42 | Sgcb | 4909 | 0.192 | 0.1852 | No |
| 43 | Pmepa1 | 5047 | 0.184 | 0.1836 | No |
| 44 | Lama1 | 5243 | 0.176 | 0.1787 | No |
| 45 | Plod2 | 5445 | 0.168 | 0.1732 | No |
| 46 | Tnfaip3 | 5472 | 0.166 | 0.1768 | No |
| 47 | Fstl1 | 5669 | 0.159 | 0.1714 | No |
| 48 | Col1a1 | 5885 | 0.149 | 0.1646 | No |
| 49 | Id2 | 5900 | 0.149 | 0.1684 | No |
| 50 | Fbln1 | 5952 | 0.145 | 0.1700 | No |
| 51 | Col4a2 | 6164 | 0.134 | 0.1630 | No |
| 52 | Slit2 | 6265 | 0.129 | 0.1617 | No |
| 53 | Magee1 | 6290 | 0.128 | 0.1643 | No |
| 54 | Col1a2 | 6305 | 0.127 | 0.1674 | No |
| 55 | Vegfa | 6551 | 0.116 | 0.1580 | No |
| 56 | Fzd8 | 6679 | 0.109 | 0.1547 | No |
| 57 | Lgals1 | 6708 | 0.108 | 0.1565 | No |
| 58 | Pvr | 6767 | 0.105 | 0.1566 | No |
| 59 | Itgb3 | 6838 | 0.102 | 0.1560 | No |
| 60 | Vegfc | 7177 | 0.086 | 0.1410 | No |
| 61 | Tagln | 7659 | 0.066 | 0.1178 | No |
| 62 | Itgav | 7861 | 0.058 | 0.1091 | No |
| 63 | Wipf1 | 8122 | 0.047 | 0.0969 | No |
| 64 | Fuca1 | 8422 | 0.033 | 0.0823 | No |
| 65 | Gja1 | 8668 | 0.021 | 0.0702 | No |
| 66 | Cdh11 | 8698 | 0.020 | 0.0692 | No |
| 67 | Gpx7 | 8777 | 0.017 | 0.0657 | No |
| 68 | Itgb5 | 9371 | -0.003 | 0.0349 | No |
| 69 | Col7a1 | 9454 | -0.007 | 0.0308 | No |
| 70 | Vcan | 9483 | -0.008 | 0.0296 | No |
| 71 | Tgfbi | 9592 | -0.013 | 0.0243 | No |
| 72 | Plaur | 9698 | -0.017 | 0.0194 | No |
| 73 | Sfrp1 | 9741 | -0.019 | 0.0177 | No |
| 74 | Tgfbr3 | 9804 | -0.022 | 0.0152 | No |
| 75 | Thy1 | 9807 | -0.022 | 0.0157 | No |
| 76 | Dab2 | 10125 | -0.035 | 0.0002 | No |
| 77 | Notch2 | 10184 | -0.038 | -0.0017 | No |
| 78 | Cxcl5 | 10318 | -0.044 | -0.0073 | No |
| 79 | Itgb1 | 10428 | -0.048 | -0.0116 | No |
| 80 | Lamc1 | 10610 | -0.055 | -0.0194 | No |
| 81 | Lrp1 | 10749 | -0.060 | -0.0248 | No |
| 82 | Matn2 | 10899 | -0.065 | -0.0306 | No |
| 83 | Sdc1 | 11237 | -0.079 | -0.0458 | No |
| 84 | Fstl3 | 11381 | -0.085 | -0.0507 | No |
| 85 | Dcn | 11508 | -0.090 | -0.0546 | No |
| 86 | Pcolce | 11649 | -0.094 | -0.0591 | No |
| 87 | Mylk | 11771 | -0.100 | -0.0624 | No |
| 88 | Sat1 | 11774 | -0.100 | -0.0595 | No |
| 89 | Tpm4 | 11899 | -0.106 | -0.0628 | No |
| 90 | Nid2 | 11961 | -0.109 | -0.0627 | No |
| 91 | Comp | 12274 | -0.121 | -0.0753 | No |
| 92 | Adam12 | 12282 | -0.122 | -0.0721 | No |
| 93 | Ntm | 12456 | -0.130 | -0.0772 | No |
| 94 | Cd44 | 12511 | -0.133 | -0.0761 | No |
| 95 | Tgm2 | 12955 | -0.153 | -0.0946 | No |
| 96 | Nt5e | 12988 | -0.154 | -0.0916 | No |
| 97 | Slc6a8 | 13040 | -0.156 | -0.0896 | No |
| 98 | Tpm1 | 13054 | -0.157 | -0.0856 | No |
| 99 | Thbs1 | 13264 | -0.167 | -0.0915 | No |
| 100 | Mcm7 | 13371 | -0.171 | -0.0919 | No |
| 101 | Pfn2 | 13647 | -0.184 | -0.1007 | No |
| 102 | Flna | 13738 | -0.189 | -0.0998 | No |
| 103 | Plod1 | 13783 | -0.191 | -0.0964 | No |
| 104 | Cald1 | 13944 | -0.198 | -0.0988 | No |
| 105 | Tfpi2 | 14086 | -0.206 | -0.1000 | No |
| 106 | Capg | 14134 | -0.206 | -0.0962 | No |
| 107 | Itga2 | 14141 | -0.206 | -0.0904 | No |
| 108 | Ppib | 14198 | -0.209 | -0.0870 | No |
| 109 | Myl9 | 15270 | -0.264 | -0.1350 | No |
| 110 | Gadd45b | 15284 | -0.265 | -0.1277 | No |
| 111 | Abi3bp | 15342 | -0.269 | -0.1226 | No |
| 112 | Col4a1 | 15631 | -0.285 | -0.1291 | No |
| 113 | Ecm1 | 15885 | -0.300 | -0.1333 | No |
| 114 | Pmp22 | 16225 | -0.322 | -0.1414 | No |
| 115 | Tgfb1 | 16242 | -0.323 | -0.1325 | No |
| 116 | Glipr1 | 16346 | -0.330 | -0.1280 | No |
| 117 | Eno2 | 17088 | -0.385 | -0.1552 | No |
| 118 | Vim | 17102 | -0.386 | -0.1443 | No |
| 119 | Tpm2 | 17110 | -0.386 | -0.1331 | No |
| 120 | Colgalt1 | 17505 | -0.418 | -0.1412 | No |
| 121 | Fas | 17610 | -0.428 | -0.1338 | No |
| 122 | Fbn1 | 17620 | -0.429 | -0.1214 | No |
| 123 | Bgn | 17744 | -0.443 | -0.1145 | No |
| 124 | Igfbp4 | 17801 | -0.450 | -0.1040 | No |
| 125 | Gem | 18073 | -0.483 | -0.1037 | No |
| 126 | Emp3 | 18102 | -0.488 | -0.0905 | No |
| 127 | Il15 | 18205 | -0.502 | -0.0808 | No |
| 128 | Fgf2 | 18279 | -0.514 | -0.0692 | No |
| 129 | Plod3 | 18421 | -0.536 | -0.0605 | No |
| 130 | Basp1 | 18466 | -0.544 | -0.0465 | No |
| 131 | Gadd45a | 18603 | -0.572 | -0.0365 | No |
| 132 | Lama2 | 18612 | -0.573 | -0.0198 | No |
| 133 | Bmp1 | 19074 | -0.747 | -0.0215 | No |
| 134 | Pdgfrb | 19278 | -1.121 | 0.0015 | No |