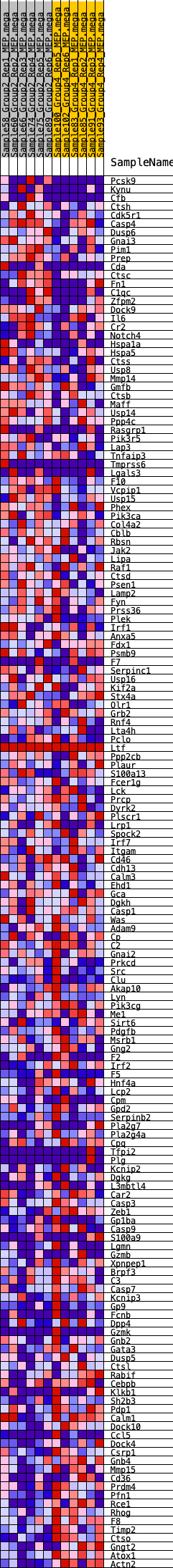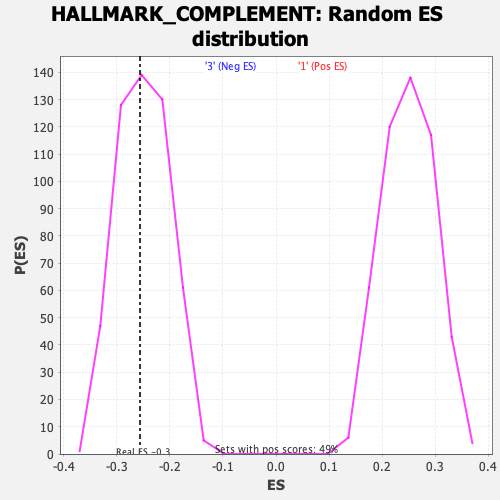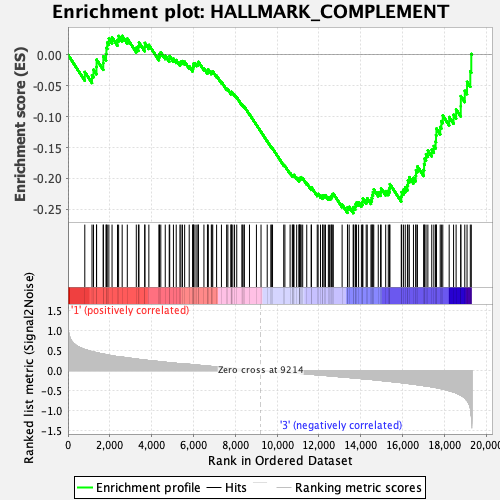
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_COMPLEMENT |
| Enrichment Score (ES) | -0.2561611 |
| Normalized Enrichment Score (NES) | -1.0284894 |
| Nominal p-value | 0.45596868 |
| FDR q-value | 0.8718021 |
| FWER p-Value | 0.976 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pcsk9 | 803 | 0.525 | -0.0278 | No |
| 2 | Kynu | 1148 | 0.473 | -0.0331 | No |
| 3 | Cfb | 1218 | 0.465 | -0.0242 | No |
| 4 | Ctsh | 1361 | 0.446 | -0.0196 | No |
| 5 | Cdk5r1 | 1368 | 0.445 | -0.0080 | No |
| 6 | Casp4 | 1685 | 0.412 | -0.0134 | No |
| 7 | Dusp6 | 1690 | 0.411 | -0.0026 | No |
| 8 | Gnai3 | 1820 | 0.398 | 0.0013 | No |
| 9 | Pim1 | 1836 | 0.396 | 0.0112 | No |
| 10 | Prep | 1876 | 0.393 | 0.0197 | No |
| 11 | Cda | 1954 | 0.385 | 0.0261 | No |
| 12 | Ctsc | 2106 | 0.369 | 0.0281 | No |
| 13 | Fn1 | 2366 | 0.347 | 0.0239 | No |
| 14 | C1qc | 2420 | 0.344 | 0.0304 | No |
| 15 | Zfpm2 | 2590 | 0.333 | 0.0305 | No |
| 16 | Dock9 | 2835 | 0.313 | 0.0262 | No |
| 17 | Il6 | 3265 | 0.286 | 0.0114 | No |
| 18 | Cr2 | 3367 | 0.279 | 0.0137 | No |
| 19 | Notch4 | 3394 | 0.277 | 0.0198 | No |
| 20 | Hspa1a | 3670 | 0.261 | 0.0124 | No |
| 21 | Hspa5 | 3676 | 0.260 | 0.0191 | No |
| 22 | Ctss | 3866 | 0.248 | 0.0159 | No |
| 23 | Usp8 | 4346 | 0.222 | -0.0031 | No |
| 24 | Mmp14 | 4376 | 0.221 | 0.0013 | No |
| 25 | Gmfb | 4437 | 0.217 | 0.0040 | No |
| 26 | Ctsb | 4650 | 0.206 | -0.0016 | No |
| 27 | Maff | 4840 | 0.195 | -0.0062 | No |
| 28 | Usp14 | 4863 | 0.193 | -0.0021 | No |
| 29 | Ppp4c | 5041 | 0.185 | -0.0064 | No |
| 30 | Rasgrp1 | 5174 | 0.178 | -0.0085 | No |
| 31 | Pik3r5 | 5352 | 0.173 | -0.0131 | No |
| 32 | Lap3 | 5400 | 0.170 | -0.0110 | No |
| 33 | Tnfaip3 | 5472 | 0.166 | -0.0103 | No |
| 34 | Tmprss6 | 5577 | 0.161 | -0.0114 | No |
| 35 | Lgals3 | 5795 | 0.153 | -0.0186 | No |
| 36 | F10 | 5959 | 0.145 | -0.0232 | No |
| 37 | Vcpip1 | 5976 | 0.144 | -0.0202 | No |
| 38 | Usp15 | 5993 | 0.143 | -0.0172 | No |
| 39 | Phex | 6008 | 0.142 | -0.0141 | No |
| 40 | Pik3ca | 6072 | 0.138 | -0.0137 | No |
| 41 | Col4a2 | 6164 | 0.134 | -0.0148 | No |
| 42 | Cblb | 6222 | 0.131 | -0.0143 | No |
| 43 | Rbsn | 6236 | 0.131 | -0.0115 | No |
| 44 | Jak2 | 6499 | 0.118 | -0.0220 | No |
| 45 | Lipa | 6667 | 0.110 | -0.0278 | No |
| 46 | Raf1 | 6690 | 0.109 | -0.0260 | No |
| 47 | Ctsd | 6702 | 0.108 | -0.0237 | No |
| 48 | Psen1 | 6852 | 0.101 | -0.0287 | No |
| 49 | Lamp2 | 6882 | 0.100 | -0.0276 | No |
| 50 | Fyn | 6920 | 0.098 | -0.0269 | No |
| 51 | Prss36 | 7105 | 0.089 | -0.0341 | No |
| 52 | Plek | 7336 | 0.080 | -0.0440 | No |
| 53 | Irf1 | 7585 | 0.069 | -0.0551 | No |
| 54 | Anxa5 | 7643 | 0.066 | -0.0563 | No |
| 55 | Fdx1 | 7794 | 0.060 | -0.0625 | No |
| 56 | Psmb9 | 7802 | 0.060 | -0.0612 | No |
| 57 | F7 | 7808 | 0.060 | -0.0599 | No |
| 58 | Serpinc1 | 7879 | 0.056 | -0.0620 | No |
| 59 | Usp16 | 7963 | 0.053 | -0.0649 | No |
| 60 | Kif2a | 8068 | 0.049 | -0.0690 | No |
| 61 | Stx4a | 8319 | 0.038 | -0.0811 | No |
| 62 | Olr1 | 8332 | 0.037 | -0.0807 | No |
| 63 | Grb2 | 8411 | 0.033 | -0.0839 | No |
| 64 | Rnf4 | 8431 | 0.032 | -0.0840 | No |
| 65 | Lta4h | 8677 | 0.020 | -0.0963 | No |
| 66 | Pclo | 9009 | 0.009 | -0.1133 | No |
| 67 | Ltf | 9229 | 0.000 | -0.1248 | No |
| 68 | Ppp2cb | 9525 | -0.010 | -0.1399 | No |
| 69 | Plaur | 9698 | -0.017 | -0.1484 | No |
| 70 | S100a13 | 9738 | -0.019 | -0.1499 | No |
| 71 | Fcer1g | 9779 | -0.021 | -0.1515 | No |
| 72 | Lck | 10314 | -0.044 | -0.1782 | No |
| 73 | Prcp | 10370 | -0.045 | -0.1798 | No |
| 74 | Dyrk2 | 10619 | -0.055 | -0.1913 | No |
| 75 | Plscr1 | 10725 | -0.059 | -0.1952 | No |
| 76 | Lrp1 | 10749 | -0.060 | -0.1948 | No |
| 77 | Spock2 | 10781 | -0.060 | -0.1948 | No |
| 78 | Irf7 | 10805 | -0.062 | -0.1943 | No |
| 79 | Itgam | 10933 | -0.066 | -0.1992 | No |
| 80 | Cd46 | 11048 | -0.072 | -0.2032 | No |
| 81 | Cdh13 | 11050 | -0.072 | -0.2013 | No |
| 82 | Calm3 | 11063 | -0.073 | -0.2000 | No |
| 83 | Ehd1 | 11086 | -0.073 | -0.1992 | No |
| 84 | Gca | 11129 | -0.075 | -0.1993 | No |
| 85 | Dgkh | 11143 | -0.076 | -0.1980 | No |
| 86 | Casp1 | 11221 | -0.079 | -0.1999 | No |
| 87 | Was | 11415 | -0.087 | -0.2076 | No |
| 88 | Adam9 | 11626 | -0.093 | -0.2161 | No |
| 89 | Cp | 11641 | -0.094 | -0.2143 | No |
| 90 | C2 | 11916 | -0.107 | -0.2257 | No |
| 91 | Gnai2 | 11959 | -0.109 | -0.2250 | No |
| 92 | Prkcd | 12073 | -0.113 | -0.2279 | No |
| 93 | Src | 12173 | -0.117 | -0.2299 | No |
| 94 | Clu | 12188 | -0.118 | -0.2275 | No |
| 95 | Akap10 | 12249 | -0.120 | -0.2274 | No |
| 96 | Lyn | 12310 | -0.123 | -0.2272 | No |
| 97 | Pik3cg | 12449 | -0.129 | -0.2309 | No |
| 98 | Me1 | 12508 | -0.132 | -0.2304 | No |
| 99 | Sirt6 | 12573 | -0.135 | -0.2301 | No |
| 100 | Pdgfb | 12606 | -0.137 | -0.2281 | No |
| 101 | Msrb1 | 12634 | -0.138 | -0.2258 | No |
| 102 | Gng2 | 12684 | -0.140 | -0.2246 | No |
| 103 | F2 | 13106 | -0.159 | -0.2424 | No |
| 104 | Irf2 | 13362 | -0.171 | -0.2511 | No |
| 105 | F5 | 13367 | -0.171 | -0.2467 | No |
| 106 | Hnf4a | 13444 | -0.175 | -0.2460 | No |
| 107 | Lcp2 | 13640 | -0.184 | -0.2512 | Yes |
| 108 | Cpm | 13643 | -0.184 | -0.2464 | Yes |
| 109 | Gpd2 | 13729 | -0.188 | -0.2458 | Yes |
| 110 | Serpinb2 | 13751 | -0.190 | -0.2418 | Yes |
| 111 | Pla2g7 | 13796 | -0.191 | -0.2389 | Yes |
| 112 | Pla2g4a | 13894 | -0.196 | -0.2387 | Yes |
| 113 | Cpq | 14028 | -0.202 | -0.2402 | Yes |
| 114 | Tfpi2 | 14086 | -0.206 | -0.2377 | Yes |
| 115 | Plg | 14096 | -0.206 | -0.2326 | Yes |
| 116 | Kcnip2 | 14257 | -0.212 | -0.2353 | Yes |
| 117 | Dgkg | 14307 | -0.214 | -0.2321 | Yes |
| 118 | L3mbtl4 | 14485 | -0.222 | -0.2354 | Yes |
| 119 | Car2 | 14527 | -0.224 | -0.2315 | Yes |
| 120 | Casp3 | 14547 | -0.226 | -0.2264 | Yes |
| 121 | Zeb1 | 14582 | -0.227 | -0.2221 | Yes |
| 122 | Gp1ba | 14619 | -0.230 | -0.2178 | Yes |
| 123 | Casp9 | 14834 | -0.242 | -0.2225 | Yes |
| 124 | S100a9 | 14945 | -0.247 | -0.2216 | Yes |
| 125 | Lgmn | 14970 | -0.249 | -0.2162 | Yes |
| 126 | Gzmb | 15189 | -0.260 | -0.2206 | Yes |
| 127 | Xpnpep1 | 15314 | -0.267 | -0.2199 | Yes |
| 128 | Brpf3 | 15375 | -0.271 | -0.2157 | Yes |
| 129 | C3 | 15391 | -0.272 | -0.2092 | Yes |
| 130 | Casp7 | 15934 | -0.303 | -0.2294 | Yes |
| 131 | Kcnip3 | 15952 | -0.304 | -0.2221 | Yes |
| 132 | Gp9 | 16037 | -0.309 | -0.2182 | Yes |
| 133 | Fcnb | 16125 | -0.316 | -0.2142 | Yes |
| 134 | Dpp4 | 16229 | -0.322 | -0.2110 | Yes |
| 135 | Gzmk | 16252 | -0.324 | -0.2034 | Yes |
| 136 | Gnb2 | 16318 | -0.328 | -0.1980 | Yes |
| 137 | Gata3 | 16517 | -0.342 | -0.1991 | Yes |
| 138 | Dusp5 | 16626 | -0.349 | -0.1954 | Yes |
| 139 | Ctsl | 16639 | -0.350 | -0.1866 | Yes |
| 140 | Rabif | 16707 | -0.355 | -0.1806 | Yes |
| 141 | Cebpb | 17006 | -0.378 | -0.1860 | Yes |
| 142 | Klkb1 | 17026 | -0.380 | -0.1768 | Yes |
| 143 | Sh2b3 | 17052 | -0.382 | -0.1678 | Yes |
| 144 | Pdp1 | 17118 | -0.387 | -0.1609 | Yes |
| 145 | Calm1 | 17206 | -0.393 | -0.1548 | Yes |
| 146 | Dock10 | 17392 | -0.409 | -0.1535 | Yes |
| 147 | Ccl5 | 17492 | -0.416 | -0.1475 | Yes |
| 148 | Dock4 | 17575 | -0.426 | -0.1404 | Yes |
| 149 | Csrp1 | 17596 | -0.427 | -0.1299 | Yes |
| 150 | Gnb4 | 17615 | -0.429 | -0.1194 | Yes |
| 151 | Mmp15 | 17796 | -0.450 | -0.1167 | Yes |
| 152 | Cd36 | 17852 | -0.455 | -0.1073 | Yes |
| 153 | Prdm4 | 17919 | -0.463 | -0.0983 | Yes |
| 154 | Pfn1 | 18225 | -0.506 | -0.1007 | Yes |
| 155 | Rce1 | 18435 | -0.539 | -0.0971 | Yes |
| 156 | Rhog | 18556 | -0.559 | -0.0883 | Yes |
| 157 | F8 | 18779 | -0.617 | -0.0833 | Yes |
| 158 | Timp2 | 18782 | -0.618 | -0.0668 | Yes |
| 159 | Ctso | 18971 | -0.684 | -0.0583 | Yes |
| 160 | Gngt2 | 19079 | -0.749 | -0.0437 | Yes |
| 161 | Atox1 | 19235 | -0.937 | -0.0267 | Yes |
| 162 | Actn2 | 19282 | -1.131 | 0.0013 | Yes |