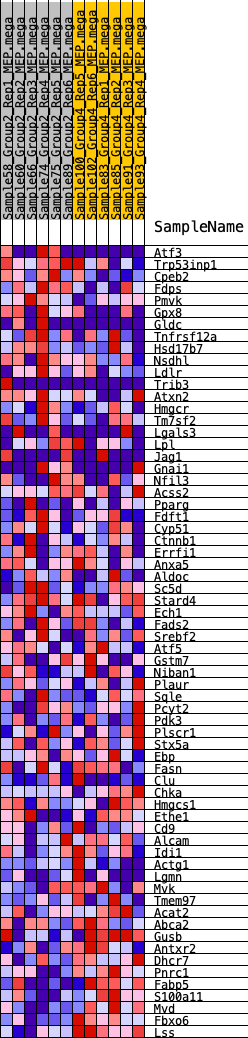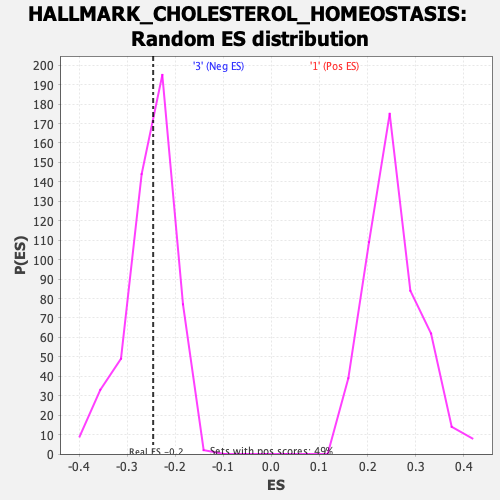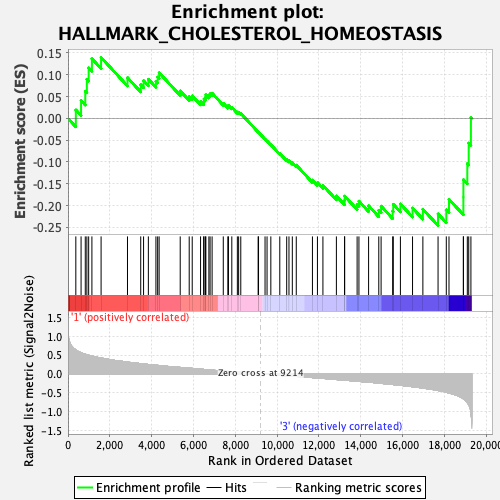
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.24592252 |
| Normalized Enrichment Score (NES) | -0.9747258 |
| Nominal p-value | 0.48133594 |
| FDR q-value | 0.858555 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Atf3 | 374 | 0.633 | 0.0193 | No |
| 2 | Trp53inp1 | 625 | 0.564 | 0.0409 | No |
| 3 | Cpeb2 | 826 | 0.521 | 0.0624 | No |
| 4 | Fdps | 902 | 0.511 | 0.0898 | No |
| 5 | Pmvk | 988 | 0.495 | 0.1157 | No |
| 6 | Gpx8 | 1143 | 0.473 | 0.1368 | No |
| 7 | Gldc | 1582 | 0.421 | 0.1398 | No |
| 8 | Tnfrsf12a | 2844 | 0.312 | 0.0934 | No |
| 9 | Hsd17b7 | 3477 | 0.272 | 0.0772 | No |
| 10 | Nsdhl | 3613 | 0.263 | 0.0864 | No |
| 11 | Ldlr | 3842 | 0.250 | 0.0898 | No |
| 12 | Trib3 | 4204 | 0.231 | 0.0852 | No |
| 13 | Atxn2 | 4287 | 0.226 | 0.0948 | No |
| 14 | Hmgcr | 4357 | 0.221 | 0.1048 | No |
| 15 | Tm7sf2 | 5365 | 0.172 | 0.0630 | No |
| 16 | Lgals3 | 5795 | 0.153 | 0.0501 | No |
| 17 | Lpl | 5939 | 0.146 | 0.0516 | No |
| 18 | Jag1 | 6338 | 0.126 | 0.0386 | No |
| 19 | Gnai1 | 6485 | 0.119 | 0.0383 | No |
| 20 | Nfil3 | 6504 | 0.118 | 0.0446 | No |
| 21 | Acss2 | 6572 | 0.115 | 0.0481 | No |
| 22 | Pparg | 6592 | 0.113 | 0.0541 | No |
| 23 | Fdft1 | 6736 | 0.106 | 0.0532 | No |
| 24 | Cyp51 | 6793 | 0.104 | 0.0566 | No |
| 25 | Ctnnb1 | 6890 | 0.099 | 0.0577 | No |
| 26 | Errfi1 | 7424 | 0.075 | 0.0347 | No |
| 27 | Anxa5 | 7643 | 0.066 | 0.0274 | No |
| 28 | Aldoc | 7672 | 0.066 | 0.0300 | No |
| 29 | Sc5d | 7832 | 0.059 | 0.0253 | No |
| 30 | Stard4 | 8097 | 0.048 | 0.0145 | No |
| 31 | Ech1 | 8158 | 0.045 | 0.0142 | No |
| 32 | Fads2 | 8261 | 0.041 | 0.0114 | No |
| 33 | Srebf2 | 9091 | 0.006 | -0.0313 | No |
| 34 | Atf5 | 9109 | 0.005 | -0.0319 | No |
| 35 | Gstm7 | 9421 | -0.006 | -0.0477 | No |
| 36 | Niban1 | 9517 | -0.010 | -0.0521 | No |
| 37 | Plaur | 9698 | -0.017 | -0.0604 | No |
| 38 | Sqle | 10126 | -0.035 | -0.0804 | No |
| 39 | Pcyt2 | 10456 | -0.049 | -0.0945 | No |
| 40 | Pdk3 | 10565 | -0.053 | -0.0969 | No |
| 41 | Plscr1 | 10725 | -0.059 | -0.1015 | No |
| 42 | Stx5a | 10914 | -0.066 | -0.1072 | No |
| 43 | Ebp | 11687 | -0.096 | -0.1414 | No |
| 44 | Fasn | 11926 | -0.108 | -0.1472 | No |
| 45 | Clu | 12188 | -0.118 | -0.1535 | No |
| 46 | Chka | 12832 | -0.147 | -0.1779 | No |
| 47 | Hmgcs1 | 13223 | -0.164 | -0.1881 | No |
| 48 | Ethe1 | 13227 | -0.164 | -0.1782 | No |
| 49 | Cd9 | 13823 | -0.193 | -0.1973 | No |
| 50 | Alcam | 13910 | -0.197 | -0.1897 | No |
| 51 | Idi1 | 14372 | -0.217 | -0.2004 | No |
| 52 | Actg1 | 14859 | -0.242 | -0.2108 | No |
| 53 | Lgmn | 14970 | -0.249 | -0.2012 | No |
| 54 | Mvk | 15523 | -0.280 | -0.2128 | No |
| 55 | Tmem97 | 15552 | -0.281 | -0.1970 | No |
| 56 | Acat2 | 15893 | -0.301 | -0.1963 | No |
| 57 | Abca2 | 16473 | -0.339 | -0.2056 | No |
| 58 | Gusb | 16967 | -0.375 | -0.2082 | No |
| 59 | Antxr2 | 17693 | -0.438 | -0.2191 | Yes |
| 60 | Dhcr7 | 18087 | -0.486 | -0.2097 | Yes |
| 61 | Pnrc1 | 18216 | -0.504 | -0.1855 | Yes |
| 62 | Fabp5 | 18904 | -0.658 | -0.1809 | Yes |
| 63 | S100a11 | 18905 | -0.658 | -0.1405 | Yes |
| 64 | Mvd | 19092 | -0.760 | -0.1036 | Yes |
| 65 | Fbxo6 | 19159 | -0.824 | -0.0565 | Yes |
| 66 | Lss | 19264 | -1.047 | 0.0022 | Yes |