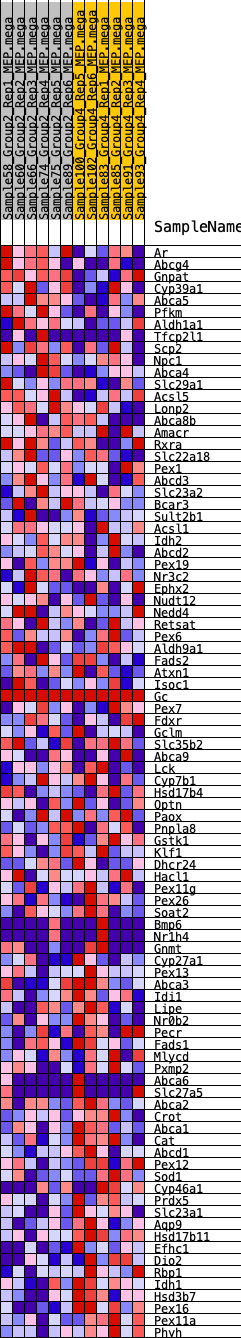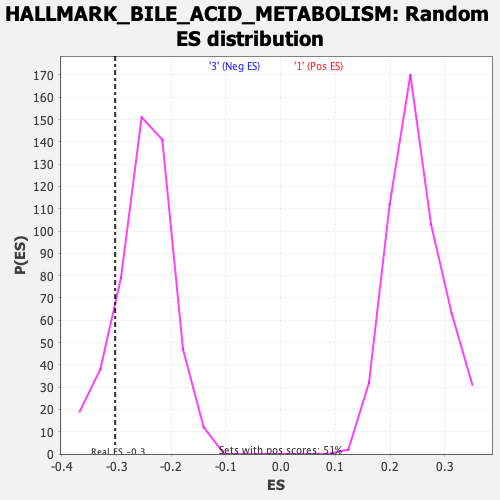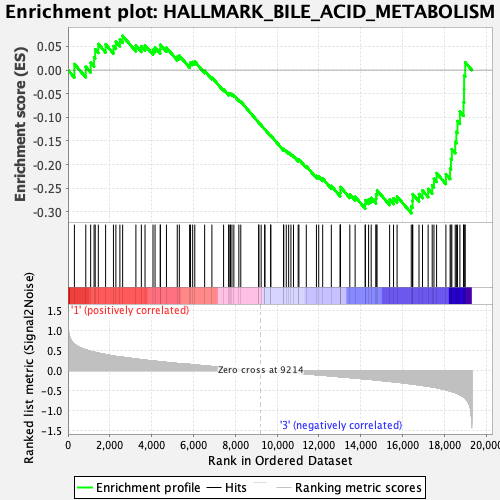
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.30263036 |
| Normalized Enrichment Score (NES) | -1.2120686 |
| Nominal p-value | 0.15811089 |
| FDR q-value | 0.6018188 |
| FWER p-Value | 0.863 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ar | 306 | 0.661 | 0.0127 | No |
| 2 | Abcg4 | 850 | 0.518 | 0.0069 | No |
| 3 | Gnpat | 1083 | 0.480 | 0.0157 | No |
| 4 | Cyp39a1 | 1247 | 0.461 | 0.0272 | No |
| 5 | Abca5 | 1301 | 0.453 | 0.0440 | No |
| 6 | Pfkm | 1450 | 0.435 | 0.0552 | No |
| 7 | Aldh1a1 | 1798 | 0.399 | 0.0544 | No |
| 8 | Tfcp2l1 | 2173 | 0.363 | 0.0507 | No |
| 9 | Scp2 | 2289 | 0.352 | 0.0600 | No |
| 10 | Npc1 | 2481 | 0.341 | 0.0648 | No |
| 11 | Abca4 | 2613 | 0.331 | 0.0723 | No |
| 12 | Slc29a1 | 3246 | 0.287 | 0.0519 | No |
| 13 | Acsl5 | 3507 | 0.271 | 0.0501 | No |
| 14 | Lonp2 | 3683 | 0.260 | 0.0523 | No |
| 15 | Abca8b | 4064 | 0.240 | 0.0429 | No |
| 16 | Amacr | 4162 | 0.234 | 0.0479 | No |
| 17 | Rxra | 4412 | 0.219 | 0.0445 | No |
| 18 | Slc22a18 | 4416 | 0.218 | 0.0538 | No |
| 19 | Pex1 | 4707 | 0.202 | 0.0475 | No |
| 20 | Abcd3 | 5224 | 0.176 | 0.0282 | No |
| 21 | Slc23a2 | 5323 | 0.174 | 0.0307 | No |
| 22 | Bcar3 | 5815 | 0.153 | 0.0117 | No |
| 23 | Sult2b1 | 5858 | 0.151 | 0.0161 | No |
| 24 | Acsl1 | 5961 | 0.145 | 0.0171 | No |
| 25 | Idh2 | 6062 | 0.139 | 0.0179 | No |
| 26 | Abcd2 | 6533 | 0.116 | -0.0016 | No |
| 27 | Pex19 | 6880 | 0.100 | -0.0152 | No |
| 28 | Nr3c2 | 7439 | 0.075 | -0.0410 | No |
| 29 | Ephx2 | 7680 | 0.065 | -0.0507 | No |
| 30 | Nudt12 | 7703 | 0.064 | -0.0491 | No |
| 31 | Nedd4 | 7770 | 0.061 | -0.0499 | No |
| 32 | Retsat | 7839 | 0.059 | -0.0508 | No |
| 33 | Pex6 | 7930 | 0.055 | -0.0532 | No |
| 34 | Aldh9a1 | 8169 | 0.045 | -0.0636 | No |
| 35 | Fads2 | 8261 | 0.041 | -0.0666 | No |
| 36 | Atxn1 | 9118 | 0.004 | -0.1109 | No |
| 37 | Isoc1 | 9123 | 0.004 | -0.1109 | No |
| 38 | Gc | 9235 | 0.000 | -0.1167 | No |
| 39 | Pex7 | 9403 | -0.005 | -0.1252 | No |
| 40 | Fdxr | 9416 | -0.005 | -0.1256 | No |
| 41 | Gclm | 9690 | -0.017 | -0.1391 | No |
| 42 | Slc35b2 | 9696 | -0.017 | -0.1386 | No |
| 43 | Abca9 | 10307 | -0.043 | -0.1684 | No |
| 44 | Lck | 10314 | -0.044 | -0.1668 | No |
| 45 | Cyp7b1 | 10436 | -0.049 | -0.1710 | No |
| 46 | Hsd17b4 | 10550 | -0.053 | -0.1746 | No |
| 47 | Optn | 10659 | -0.057 | -0.1778 | No |
| 48 | Paox | 10782 | -0.060 | -0.1815 | No |
| 49 | Pnpla8 | 11005 | -0.070 | -0.1900 | No |
| 50 | Gstk1 | 11041 | -0.072 | -0.1888 | No |
| 51 | Klf1 | 11393 | -0.086 | -0.2033 | No |
| 52 | Dhcr24 | 11878 | -0.105 | -0.2239 | No |
| 53 | Hacl1 | 11987 | -0.110 | -0.2248 | No |
| 54 | Pex11g | 12177 | -0.117 | -0.2295 | No |
| 55 | Pex26 | 12590 | -0.136 | -0.2451 | No |
| 56 | Soat2 | 13010 | -0.156 | -0.2601 | No |
| 57 | Bmp6 | 13025 | -0.156 | -0.2541 | No |
| 58 | Nr1h4 | 13026 | -0.156 | -0.2473 | No |
| 59 | Gnmt | 13474 | -0.176 | -0.2630 | No |
| 60 | Cyp27a1 | 13728 | -0.188 | -0.2680 | No |
| 61 | Pex13 | 14208 | -0.210 | -0.2838 | No |
| 62 | Abca3 | 14218 | -0.210 | -0.2752 | No |
| 63 | Idi1 | 14372 | -0.217 | -0.2737 | No |
| 64 | Lipe | 14496 | -0.223 | -0.2705 | No |
| 65 | Nr0b2 | 14721 | -0.235 | -0.2720 | No |
| 66 | Pecr | 14733 | -0.235 | -0.2623 | No |
| 67 | Fads1 | 14777 | -0.238 | -0.2543 | No |
| 68 | Mlycd | 15378 | -0.271 | -0.2738 | No |
| 69 | Pxmp2 | 15564 | -0.281 | -0.2712 | No |
| 70 | Abca6 | 15734 | -0.290 | -0.2674 | No |
| 71 | Slc27a5 | 16412 | -0.333 | -0.2882 | Yes |
| 72 | Abca2 | 16473 | -0.339 | -0.2766 | Yes |
| 73 | Crot | 16484 | -0.340 | -0.2624 | Yes |
| 74 | Abca1 | 16782 | -0.361 | -0.2622 | Yes |
| 75 | Cat | 16949 | -0.374 | -0.2547 | Yes |
| 76 | Abcd1 | 17219 | -0.395 | -0.2516 | Yes |
| 77 | Pex12 | 17413 | -0.411 | -0.2438 | Yes |
| 78 | Sod1 | 17497 | -0.417 | -0.2301 | Yes |
| 79 | Cyp46a1 | 17623 | -0.429 | -0.2180 | Yes |
| 80 | Prdx5 | 18068 | -0.483 | -0.2201 | Yes |
| 81 | Slc23a1 | 18270 | -0.512 | -0.2084 | Yes |
| 82 | Aqp9 | 18311 | -0.519 | -0.1880 | Yes |
| 83 | Hsd17b11 | 18352 | -0.525 | -0.1673 | Yes |
| 84 | Efhc1 | 18514 | -0.552 | -0.1518 | Yes |
| 85 | Dio2 | 18570 | -0.563 | -0.1302 | Yes |
| 86 | Rbp1 | 18621 | -0.575 | -0.1079 | Yes |
| 87 | Idh1 | 18737 | -0.604 | -0.0877 | Yes |
| 88 | Hsd3b7 | 18912 | -0.661 | -0.0681 | Yes |
| 89 | Pex16 | 18935 | -0.670 | -0.0402 | Yes |
| 90 | Pex11a | 18940 | -0.672 | -0.0113 | Yes |
| 91 | Phyh | 18993 | -0.700 | 0.0163 | Yes |