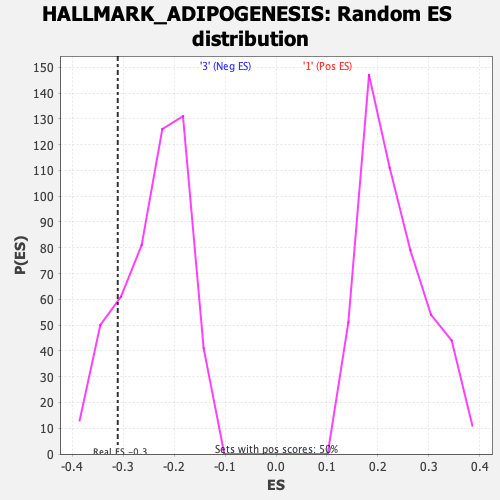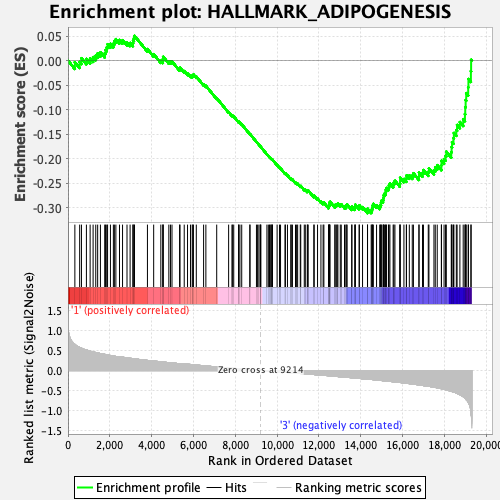
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group4.MEP.mega_Pheno.cls #Group2_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_ADIPOGENESIS |
| Enrichment Score (ES) | -0.31102458 |
| Normalized Enrichment Score (NES) | -1.3001587 |
| Nominal p-value | 0.16699801 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.769 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Lifr | 328 | 0.654 | -0.0032 | No |
| 2 | Nkiras1 | 558 | 0.578 | -0.0028 | No |
| 3 | Sspn | 643 | 0.559 | 0.0047 | No |
| 4 | Mccc1 | 880 | 0.513 | 0.0034 | No |
| 5 | Ppp1r15b | 1058 | 0.484 | 0.0044 | No |
| 6 | Atl2 | 1200 | 0.468 | 0.0071 | No |
| 7 | Cdkn2c | 1327 | 0.450 | 0.0101 | No |
| 8 | Ptcd3 | 1418 | 0.439 | 0.0148 | No |
| 9 | Gpx3 | 1550 | 0.422 | 0.0169 | No |
| 10 | Gphn | 1756 | 0.405 | 0.0149 | No |
| 11 | Cd302 | 1791 | 0.400 | 0.0216 | No |
| 12 | Aco2 | 1852 | 0.395 | 0.0269 | No |
| 13 | Adcy6 | 1892 | 0.392 | 0.0333 | No |
| 14 | Chuk | 2029 | 0.377 | 0.0342 | No |
| 15 | Mgll | 2169 | 0.363 | 0.0347 | No |
| 16 | Lama4 | 2217 | 0.359 | 0.0399 | No |
| 17 | Scp2 | 2289 | 0.352 | 0.0437 | No |
| 18 | Agpat3 | 2462 | 0.343 | 0.0420 | No |
| 19 | Ubqln1 | 2608 | 0.331 | 0.0415 | No |
| 20 | Plin2 | 2821 | 0.314 | 0.0371 | No |
| 21 | Ak2 | 2969 | 0.304 | 0.0359 | No |
| 22 | Immt | 3103 | 0.297 | 0.0353 | No |
| 23 | Dld | 3113 | 0.297 | 0.0412 | No |
| 24 | Bckdha | 3152 | 0.295 | 0.0455 | No |
| 25 | Gpat4 | 3180 | 0.293 | 0.0504 | No |
| 26 | Sorbs1 | 3798 | 0.252 | 0.0235 | No |
| 27 | Dnajc15 | 4096 | 0.238 | 0.0130 | No |
| 28 | Ndufa5 | 4438 | 0.217 | -0.0002 | No |
| 29 | Acly | 4534 | 0.213 | -0.0006 | No |
| 30 | Ppm1b | 4543 | 0.213 | 0.0035 | No |
| 31 | Dgat1 | 4550 | 0.213 | 0.0078 | No |
| 32 | Uqcrq | 4821 | 0.196 | -0.0022 | No |
| 33 | Dhrs7b | 4899 | 0.192 | -0.0021 | No |
| 34 | Tank | 4972 | 0.188 | -0.0018 | No |
| 35 | Pdcd4 | 5335 | 0.173 | -0.0171 | No |
| 36 | Acox1 | 5355 | 0.172 | -0.0144 | No |
| 37 | Cmpk1 | 5557 | 0.162 | -0.0214 | No |
| 38 | Acaa2 | 5715 | 0.157 | -0.0263 | No |
| 39 | Ddt | 5856 | 0.151 | -0.0304 | No |
| 40 | Lpl | 5939 | 0.146 | -0.0316 | No |
| 41 | Uqcr11 | 5971 | 0.144 | -0.0301 | No |
| 42 | Acadm | 5996 | 0.143 | -0.0283 | No |
| 43 | Acads | 6134 | 0.135 | -0.0326 | No |
| 44 | Coq9 | 6483 | 0.119 | -0.0482 | No |
| 45 | Pparg | 6592 | 0.113 | -0.0515 | No |
| 46 | Coq3 | 7110 | 0.089 | -0.0766 | No |
| 47 | Ephx2 | 7680 | 0.065 | -0.1050 | No |
| 48 | Retsat | 7839 | 0.059 | -0.1120 | No |
| 49 | Cavin1 | 7872 | 0.057 | -0.1124 | No |
| 50 | Dlat | 7924 | 0.055 | -0.1139 | No |
| 51 | Ech1 | 8158 | 0.045 | -0.1251 | No |
| 52 | Ywhag | 8160 | 0.045 | -0.1242 | No |
| 53 | Uqcr10 | 8219 | 0.043 | -0.1263 | No |
| 54 | Slc25a1 | 8306 | 0.039 | -0.1300 | No |
| 55 | Ucp2 | 8694 | 0.020 | -0.1498 | No |
| 56 | Dram2 | 8701 | 0.020 | -0.1497 | No |
| 57 | Itsn1 | 9010 | 0.008 | -0.1656 | No |
| 58 | Mrpl15 | 9065 | 0.007 | -0.1683 | No |
| 59 | Cyp4b1 | 9128 | 0.004 | -0.1715 | No |
| 60 | Mtch2 | 9191 | 0.001 | -0.1747 | No |
| 61 | Uqcrc1 | 9203 | 0.000 | -0.1753 | No |
| 62 | Cmbl | 9222 | 0.000 | -0.1762 | No |
| 63 | Rreb1 | 9511 | -0.009 | -0.1911 | No |
| 64 | Mgst3 | 9579 | -0.012 | -0.1943 | No |
| 65 | Ccng2 | 9635 | -0.015 | -0.1969 | No |
| 66 | Sqor | 9666 | -0.016 | -0.1981 | No |
| 67 | Gpam | 9707 | -0.018 | -0.1998 | No |
| 68 | Slc1a5 | 9748 | -0.019 | -0.2015 | No |
| 69 | Phldb1 | 9753 | -0.020 | -0.2013 | No |
| 70 | Dbt | 9777 | -0.021 | -0.2020 | No |
| 71 | Esrra | 9998 | -0.030 | -0.2129 | No |
| 72 | Aldoa | 10113 | -0.034 | -0.2181 | No |
| 73 | Lpcat3 | 10162 | -0.036 | -0.2199 | No |
| 74 | Ptger3 | 10375 | -0.046 | -0.2300 | No |
| 75 | Preb | 10395 | -0.046 | -0.2300 | No |
| 76 | G3bp2 | 10493 | -0.050 | -0.2340 | No |
| 77 | Riok3 | 10650 | -0.057 | -0.2409 | No |
| 78 | Aplp2 | 10676 | -0.058 | -0.2410 | No |
| 79 | Elovl6 | 10731 | -0.060 | -0.2425 | No |
| 80 | Ghitm | 10877 | -0.064 | -0.2487 | No |
| 81 | Bcl2l13 | 10931 | -0.066 | -0.2501 | No |
| 82 | Hadh | 10980 | -0.069 | -0.2511 | No |
| 83 | Apoe | 11105 | -0.074 | -0.2560 | No |
| 84 | Hspb8 | 11128 | -0.075 | -0.2556 | No |
| 85 | Stom | 11297 | -0.082 | -0.2626 | No |
| 86 | Baz2a | 11346 | -0.084 | -0.2633 | No |
| 87 | Tkt | 11440 | -0.087 | -0.2663 | No |
| 88 | Map4k3 | 11457 | -0.088 | -0.2653 | No |
| 89 | Sult1a1 | 11482 | -0.089 | -0.2646 | No |
| 90 | Cpt2 | 11753 | -0.099 | -0.2766 | No |
| 91 | Mylk | 11771 | -0.100 | -0.2754 | No |
| 92 | Dhrs7 | 11931 | -0.108 | -0.2814 | No |
| 93 | Stat5a | 12093 | -0.114 | -0.2874 | No |
| 94 | Fzd4 | 12197 | -0.118 | -0.2902 | No |
| 95 | Gbe1 | 12234 | -0.120 | -0.2896 | No |
| 96 | Nabp1 | 12457 | -0.130 | -0.2984 | No |
| 97 | Cd151 | 12461 | -0.130 | -0.2958 | No |
| 98 | Cavin2 | 12490 | -0.132 | -0.2944 | No |
| 99 | Pfkfb3 | 12503 | -0.132 | -0.2922 | No |
| 100 | Me1 | 12508 | -0.132 | -0.2896 | No |
| 101 | Slc25a10 | 12528 | -0.133 | -0.2877 | No |
| 102 | Idh3a | 12759 | -0.144 | -0.2967 | No |
| 103 | Pfkl | 12769 | -0.144 | -0.2941 | No |
| 104 | Atp1b3 | 12827 | -0.147 | -0.2939 | No |
| 105 | Araf | 12869 | -0.149 | -0.2929 | No |
| 106 | Coq5 | 12904 | -0.151 | -0.2914 | No |
| 107 | Tob1 | 13018 | -0.156 | -0.2940 | No |
| 108 | Elmod3 | 13064 | -0.157 | -0.2930 | No |
| 109 | Mtarc2 | 13220 | -0.164 | -0.2976 | No |
| 110 | Samm50 | 13290 | -0.168 | -0.2976 | No |
| 111 | Acadl | 13302 | -0.168 | -0.2946 | No |
| 112 | Ifngr1 | 13346 | -0.170 | -0.2932 | No |
| 113 | Slc5a6 | 13564 | -0.180 | -0.3007 | No |
| 114 | Dnajb9 | 13576 | -0.181 | -0.2974 | No |
| 115 | Cyc1 | 13712 | -0.187 | -0.3005 | No |
| 116 | Gpd2 | 13729 | -0.188 | -0.2973 | No |
| 117 | Pex14 | 13737 | -0.189 | -0.2936 | No |
| 118 | Scarb1 | 13920 | -0.197 | -0.2989 | No |
| 119 | Sowahc | 13929 | -0.198 | -0.2951 | No |
| 120 | Rab34 | 14083 | -0.206 | -0.2987 | No |
| 121 | Suclg1 | 14319 | -0.214 | -0.3064 | Yes |
| 122 | Pemt | 14322 | -0.215 | -0.3020 | Yes |
| 123 | Lipe | 14496 | -0.223 | -0.3062 | Yes |
| 124 | Bcl6 | 14528 | -0.224 | -0.3031 | Yes |
| 125 | Reep5 | 14551 | -0.226 | -0.2994 | Yes |
| 126 | Rmdn3 | 14565 | -0.227 | -0.2952 | Yes |
| 127 | Decr1 | 14598 | -0.228 | -0.2920 | Yes |
| 128 | Rnf11 | 14753 | -0.237 | -0.2950 | Yes |
| 129 | Aldh2 | 14904 | -0.245 | -0.2976 | Yes |
| 130 | Adipor2 | 14939 | -0.247 | -0.2941 | Yes |
| 131 | Sdhb | 14966 | -0.249 | -0.2902 | Yes |
| 132 | Omd | 14982 | -0.250 | -0.2856 | Yes |
| 133 | Gpx4 | 15060 | -0.254 | -0.2842 | Yes |
| 134 | Miga2 | 15083 | -0.256 | -0.2799 | Yes |
| 135 | Rtn3 | 15089 | -0.256 | -0.2747 | Yes |
| 136 | Hibch | 15128 | -0.257 | -0.2712 | Yes |
| 137 | Arl4a | 15185 | -0.260 | -0.2686 | Yes |
| 138 | Angpt1 | 15197 | -0.260 | -0.2636 | Yes |
| 139 | Cs | 15231 | -0.262 | -0.2597 | Yes |
| 140 | Aifm1 | 15323 | -0.268 | -0.2588 | Yes |
| 141 | Prdx3 | 15345 | -0.269 | -0.2541 | Yes |
| 142 | C3 | 15391 | -0.272 | -0.2507 | Yes |
| 143 | Slc27a1 | 15546 | -0.281 | -0.2527 | Yes |
| 144 | Angptl4 | 15580 | -0.282 | -0.2484 | Yes |
| 145 | Col4a1 | 15631 | -0.285 | -0.2449 | Yes |
| 146 | Qdpr | 15864 | -0.299 | -0.2507 | Yes |
| 147 | Crat | 15879 | -0.300 | -0.2450 | Yes |
| 148 | Cox8a | 15880 | -0.300 | -0.2386 | Yes |
| 149 | Ndufs3 | 16061 | -0.311 | -0.2414 | Yes |
| 150 | Por | 16176 | -0.318 | -0.2405 | Yes |
| 151 | Mdh2 | 16185 | -0.319 | -0.2341 | Yes |
| 152 | Abcb8 | 16319 | -0.328 | -0.2341 | Yes |
| 153 | Itih5 | 16460 | -0.338 | -0.2342 | Yes |
| 154 | Nmt1 | 16515 | -0.342 | -0.2297 | Yes |
| 155 | Jagn1 | 16780 | -0.360 | -0.2358 | Yes |
| 156 | Abca1 | 16782 | -0.361 | -0.2282 | Yes |
| 157 | Cat | 16949 | -0.374 | -0.2289 | Yes |
| 158 | Pgm1 | 16995 | -0.377 | -0.2232 | Yes |
| 159 | Etfb | 17225 | -0.395 | -0.2267 | Yes |
| 160 | Grpel1 | 17257 | -0.397 | -0.2198 | Yes |
| 161 | Sod1 | 17497 | -0.417 | -0.2234 | Yes |
| 162 | Echs1 | 17559 | -0.424 | -0.2176 | Yes |
| 163 | Sdhc | 17654 | -0.433 | -0.2132 | Yes |
| 164 | Cd36 | 17852 | -0.455 | -0.2138 | Yes |
| 165 | Uck1 | 17856 | -0.455 | -0.2042 | Yes |
| 166 | Esyt1 | 17980 | -0.469 | -0.2006 | Yes |
| 167 | Taldo1 | 18052 | -0.481 | -0.1941 | Yes |
| 168 | Dhcr7 | 18087 | -0.486 | -0.1855 | Yes |
| 169 | Slc19a1 | 18325 | -0.522 | -0.1867 | Yes |
| 170 | Pim3 | 18341 | -0.523 | -0.1763 | Yes |
| 171 | Ltc4s | 18363 | -0.527 | -0.1662 | Yes |
| 172 | Ubc | 18431 | -0.538 | -0.1582 | Yes |
| 173 | Chchd10 | 18450 | -0.541 | -0.1476 | Yes |
| 174 | Reep6 | 18566 | -0.562 | -0.1416 | Yes |
| 175 | Gadd45a | 18603 | -0.572 | -0.1312 | Yes |
| 176 | Idh1 | 18737 | -0.604 | -0.1253 | Yes |
| 177 | Cox6a1 | 18895 | -0.655 | -0.1195 | Yes |
| 178 | Cox7b | 18979 | -0.691 | -0.1091 | Yes |
| 179 | Phyh | 18993 | -0.700 | -0.0948 | Yes |
| 180 | Fah | 19012 | -0.709 | -0.0806 | Yes |
| 181 | Vegfb | 19040 | -0.731 | -0.0664 | Yes |
| 182 | Idh3g | 19134 | -0.791 | -0.0543 | Yes |
| 183 | Ndufab1 | 19145 | -0.809 | -0.0376 | Yes |
| 184 | Tst | 19263 | -1.042 | -0.0214 | Yes |
| 185 | Ndufb7 | 19274 | -1.109 | 0.0017 | Yes |