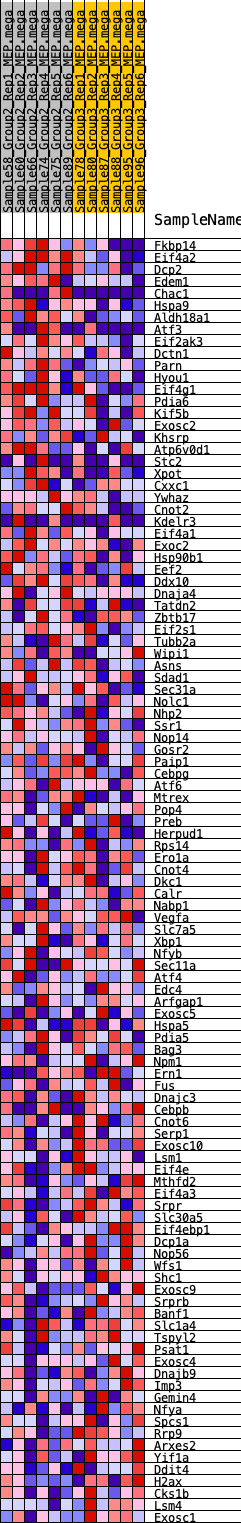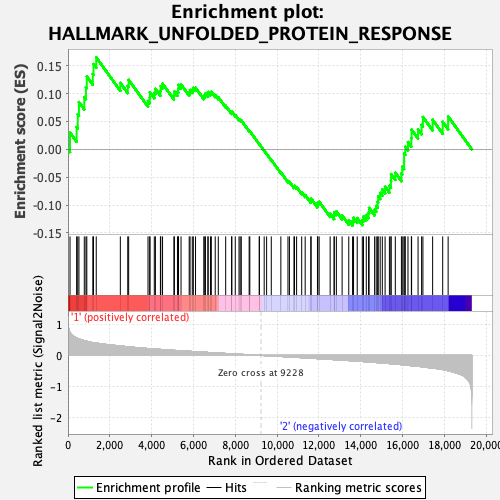
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.16483729 |
| Normalized Enrichment Score (NES) | 0.6984368 |
| Nominal p-value | 0.8510204 |
| FDR q-value | 1.0 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Fkbp14 | 96 | 0.745 | 0.0300 | Yes |
| 2 | Eif4a2 | 412 | 0.562 | 0.0399 | Yes |
| 3 | Dcp2 | 466 | 0.544 | 0.0627 | Yes |
| 4 | Edem1 | 523 | 0.530 | 0.0847 | Yes |
| 5 | Chac1 | 779 | 0.477 | 0.0938 | Yes |
| 6 | Hspa9 | 858 | 0.463 | 0.1115 | Yes |
| 7 | Aldh18a1 | 899 | 0.454 | 0.1307 | Yes |
| 8 | Atf3 | 1187 | 0.414 | 0.1352 | Yes |
| 9 | Eif2ak3 | 1219 | 0.411 | 0.1529 | Yes |
| 10 | Dctn1 | 1350 | 0.398 | 0.1648 | Yes |
| 11 | Parn | 2502 | 0.304 | 0.1192 | No |
| 12 | Hyou1 | 2857 | 0.279 | 0.1138 | No |
| 13 | Eif4g1 | 2900 | 0.276 | 0.1246 | No |
| 14 | Pdia6 | 3826 | 0.221 | 0.0868 | No |
| 15 | Kif5b | 3910 | 0.216 | 0.0927 | No |
| 16 | Exosc2 | 3917 | 0.216 | 0.1025 | No |
| 17 | Khsrp | 4126 | 0.208 | 0.1014 | No |
| 18 | Atp6v0d1 | 4179 | 0.206 | 0.1084 | No |
| 19 | Stc2 | 4418 | 0.194 | 0.1051 | No |
| 20 | Xpot | 4435 | 0.192 | 0.1133 | No |
| 21 | Cxxc1 | 4520 | 0.188 | 0.1177 | No |
| 22 | Ywhaz | 5064 | 0.164 | 0.0971 | No |
| 23 | Cnot2 | 5086 | 0.162 | 0.1036 | No |
| 24 | Kdelr3 | 5234 | 0.157 | 0.1034 | No |
| 25 | Eif4a1 | 5273 | 0.156 | 0.1087 | No |
| 26 | Exoc2 | 5282 | 0.155 | 0.1156 | No |
| 27 | Hsp90b1 | 5404 | 0.150 | 0.1163 | No |
| 28 | Eef2 | 5794 | 0.134 | 0.1024 | No |
| 29 | Ddx10 | 5835 | 0.132 | 0.1065 | No |
| 30 | Dnaja4 | 5942 | 0.127 | 0.1069 | No |
| 31 | Tatdn2 | 5992 | 0.125 | 0.1102 | No |
| 32 | Zbtb17 | 6097 | 0.120 | 0.1104 | No |
| 33 | Eif2s1 | 6492 | 0.103 | 0.0948 | No |
| 34 | Tubb2a | 6538 | 0.102 | 0.0972 | No |
| 35 | Wipi1 | 6571 | 0.100 | 0.1003 | No |
| 36 | Asns | 6693 | 0.096 | 0.0985 | No |
| 37 | Sdad1 | 6696 | 0.096 | 0.1029 | No |
| 38 | Sec31a | 6811 | 0.091 | 0.1012 | No |
| 39 | Nolc1 | 6853 | 0.089 | 0.1033 | No |
| 40 | Nhp2 | 7039 | 0.082 | 0.0975 | No |
| 41 | Ssr1 | 7186 | 0.076 | 0.0934 | No |
| 42 | Nop14 | 7539 | 0.063 | 0.0781 | No |
| 43 | Gosr2 | 7821 | 0.053 | 0.0659 | No |
| 44 | Paip1 | 7835 | 0.053 | 0.0677 | No |
| 45 | Cebpg | 7997 | 0.046 | 0.0615 | No |
| 46 | Atf6 | 8177 | 0.039 | 0.0540 | No |
| 47 | Mtrex | 8232 | 0.037 | 0.0529 | No |
| 48 | Pop4 | 8285 | 0.035 | 0.0518 | No |
| 49 | Preb | 8661 | 0.021 | 0.0333 | No |
| 50 | Herpud1 | 8693 | 0.020 | 0.0326 | No |
| 51 | Rps14 | 9145 | 0.004 | 0.0093 | No |
| 52 | Ero1a | 9150 | 0.003 | 0.0092 | No |
| 53 | Cnot4 | 9380 | -0.003 | -0.0025 | No |
| 54 | Dkc1 | 9496 | -0.007 | -0.0082 | No |
| 55 | Calr | 9719 | -0.015 | -0.0190 | No |
| 56 | Nabp1 | 10177 | -0.032 | -0.0413 | No |
| 57 | Vegfa | 10517 | -0.046 | -0.0568 | No |
| 58 | Slc7a5 | 10587 | -0.048 | -0.0582 | No |
| 59 | Xbp1 | 10804 | -0.056 | -0.0668 | No |
| 60 | Nfyb | 10830 | -0.057 | -0.0654 | No |
| 61 | Sec11a | 10930 | -0.061 | -0.0677 | No |
| 62 | Atf4 | 11174 | -0.071 | -0.0770 | No |
| 63 | Edc4 | 11340 | -0.077 | -0.0820 | No |
| 64 | Arfgap1 | 11610 | -0.087 | -0.0919 | No |
| 65 | Exosc5 | 11623 | -0.087 | -0.0885 | No |
| 66 | Hspa5 | 11927 | -0.101 | -0.0995 | No |
| 67 | Pdia5 | 11945 | -0.102 | -0.0956 | No |
| 68 | Bag3 | 12008 | -0.104 | -0.0939 | No |
| 69 | Npm1 | 12534 | -0.127 | -0.1154 | No |
| 70 | Ern1 | 12723 | -0.135 | -0.1188 | No |
| 71 | Fus | 12730 | -0.136 | -0.1127 | No |
| 72 | Dnajc3 | 12831 | -0.139 | -0.1114 | No |
| 73 | Cebpb | 13106 | -0.152 | -0.1185 | No |
| 74 | Cnot6 | 13424 | -0.165 | -0.1273 | No |
| 75 | Serp1 | 13606 | -0.173 | -0.1286 | No |
| 76 | Exosc10 | 13652 | -0.176 | -0.1227 | No |
| 77 | Lsm1 | 13828 | -0.182 | -0.1233 | No |
| 78 | Eif4e | 14071 | -0.194 | -0.1267 | No |
| 79 | Mthfd2 | 14128 | -0.197 | -0.1204 | No |
| 80 | Eif4a3 | 14262 | -0.204 | -0.1177 | No |
| 81 | Srpr | 14371 | -0.210 | -0.1135 | No |
| 82 | Slc30a5 | 14396 | -0.211 | -0.1049 | No |
| 83 | Eif4ebp1 | 14660 | -0.223 | -0.1081 | No |
| 84 | Dcp1a | 14736 | -0.228 | -0.1013 | No |
| 85 | Nop56 | 14805 | -0.231 | -0.0940 | No |
| 86 | Wfs1 | 14832 | -0.233 | -0.0844 | No |
| 87 | Shc1 | 14926 | -0.238 | -0.0781 | No |
| 88 | Exosc9 | 15028 | -0.243 | -0.0719 | No |
| 89 | Srprb | 15161 | -0.250 | -0.0670 | No |
| 90 | Banf1 | 15367 | -0.262 | -0.0654 | No |
| 91 | Slc1a4 | 15440 | -0.266 | -0.0567 | No |
| 92 | Tspyl2 | 15447 | -0.266 | -0.0445 | No |
| 93 | Psat1 | 15649 | -0.277 | -0.0419 | No |
| 94 | Exosc4 | 15941 | -0.297 | -0.0431 | No |
| 95 | Dnajb9 | 15981 | -0.300 | -0.0311 | No |
| 96 | Imp3 | 16068 | -0.306 | -0.0212 | No |
| 97 | Gemin4 | 16071 | -0.306 | -0.0069 | No |
| 98 | Nfya | 16128 | -0.310 | 0.0048 | No |
| 99 | Spcs1 | 16255 | -0.318 | 0.0132 | No |
| 100 | Rrp9 | 16417 | -0.329 | 0.0202 | No |
| 101 | Arxes2 | 16429 | -0.330 | 0.0352 | No |
| 102 | Yif1a | 16737 | -0.350 | 0.0356 | No |
| 103 | Ddit4 | 16906 | -0.364 | 0.0439 | No |
| 104 | H2ax | 16969 | -0.368 | 0.0580 | No |
| 105 | Cks1b | 17434 | -0.408 | 0.0530 | No |
| 106 | Lsm4 | 17915 | -0.454 | 0.0493 | No |
| 107 | Exosc1 | 18175 | -0.493 | 0.0590 | No |