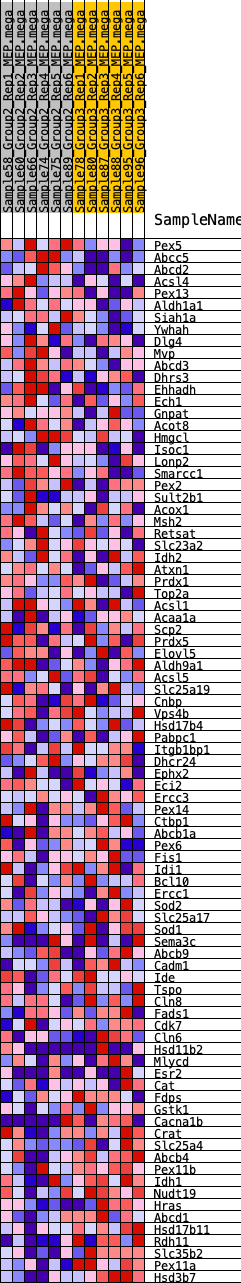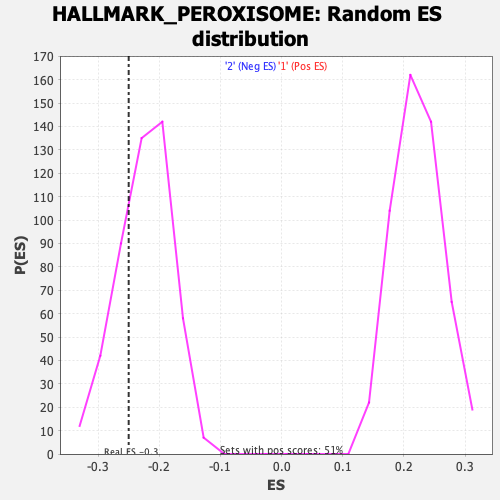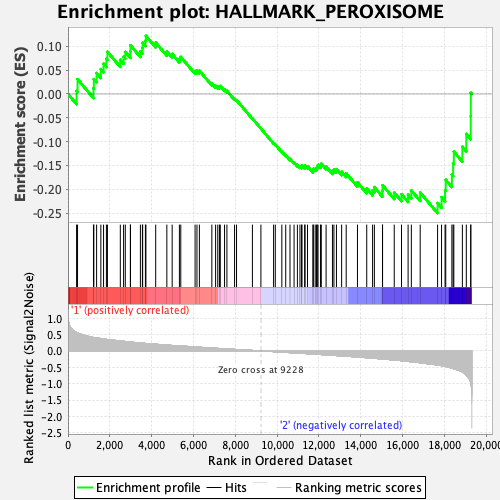
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.25019488 |
| Normalized Enrichment Score (NES) | -1.1216722 |
| Nominal p-value | 0.2674897 |
| FDR q-value | 0.8000654 |
| FWER p-Value | 0.966 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 411 | 0.562 | 0.0064 | No |
| 2 | Abcc5 | 456 | 0.546 | 0.0311 | No |
| 3 | Abcd2 | 1218 | 0.411 | 0.0119 | No |
| 4 | Acsl4 | 1236 | 0.410 | 0.0313 | No |
| 5 | Pex13 | 1368 | 0.396 | 0.0440 | No |
| 6 | Aldh1a1 | 1571 | 0.379 | 0.0523 | No |
| 7 | Siah1a | 1700 | 0.365 | 0.0636 | No |
| 8 | Ywhah | 1836 | 0.353 | 0.0741 | No |
| 9 | Dlg4 | 1885 | 0.349 | 0.0888 | No |
| 10 | Mvp | 2501 | 0.304 | 0.0719 | No |
| 11 | Abcd3 | 2658 | 0.294 | 0.0783 | No |
| 12 | Dhrs3 | 2739 | 0.288 | 0.0884 | No |
| 13 | Ehhadh | 2982 | 0.271 | 0.0892 | No |
| 14 | Ech1 | 2987 | 0.271 | 0.1024 | No |
| 15 | Gnpat | 3461 | 0.241 | 0.0897 | No |
| 16 | Acot8 | 3557 | 0.236 | 0.0965 | No |
| 17 | Hmgcl | 3570 | 0.236 | 0.1075 | No |
| 18 | Isoc1 | 3701 | 0.229 | 0.1120 | No |
| 19 | Lonp2 | 3725 | 0.227 | 0.1221 | No |
| 20 | Smarcc1 | 4193 | 0.205 | 0.1079 | No |
| 21 | Pex2 | 4726 | 0.178 | 0.0891 | No |
| 22 | Sult2b1 | 4980 | 0.167 | 0.0842 | No |
| 23 | Acox1 | 5326 | 0.154 | 0.0738 | No |
| 24 | Msh2 | 5392 | 0.151 | 0.0779 | No |
| 25 | Retsat | 6076 | 0.120 | 0.0483 | No |
| 26 | Slc23a2 | 6166 | 0.117 | 0.0495 | No |
| 27 | Idh2 | 6282 | 0.112 | 0.0490 | No |
| 28 | Atxn1 | 6878 | 0.089 | 0.0224 | No |
| 29 | Prdx1 | 7057 | 0.081 | 0.0172 | No |
| 30 | Top2a | 7161 | 0.077 | 0.0156 | No |
| 31 | Acsl1 | 7241 | 0.074 | 0.0152 | No |
| 32 | Acaa1a | 7281 | 0.073 | 0.0167 | No |
| 33 | Scp2 | 7485 | 0.065 | 0.0094 | No |
| 34 | Prdx5 | 7598 | 0.060 | 0.0066 | No |
| 35 | Elovl5 | 7964 | 0.047 | -0.0101 | No |
| 36 | Aldh9a1 | 8058 | 0.043 | -0.0128 | No |
| 37 | Acsl5 | 8817 | 0.015 | -0.0514 | No |
| 38 | Slc25a19 | 9223 | 0.000 | -0.0725 | No |
| 39 | Cnbp | 9830 | -0.019 | -0.1031 | No |
| 40 | Vps4b | 9911 | -0.022 | -0.1062 | No |
| 41 | Hsd17b4 | 10224 | -0.034 | -0.1208 | No |
| 42 | Pabpc1 | 10410 | -0.041 | -0.1283 | No |
| 43 | Itgb1bp1 | 10614 | -0.050 | -0.1365 | No |
| 44 | Dhcr24 | 10809 | -0.056 | -0.1438 | No |
| 45 | Ephx2 | 10968 | -0.062 | -0.1489 | No |
| 46 | Eci2 | 11090 | -0.068 | -0.1518 | No |
| 47 | Ercc3 | 11168 | -0.071 | -0.1523 | No |
| 48 | Pex14 | 11186 | -0.071 | -0.1497 | No |
| 49 | Ctbp1 | 11315 | -0.076 | -0.1526 | No |
| 50 | Abcb1a | 11334 | -0.077 | -0.1497 | No |
| 51 | Pex6 | 11452 | -0.081 | -0.1518 | No |
| 52 | Fis1 | 11708 | -0.091 | -0.1606 | No |
| 53 | Idi1 | 11730 | -0.092 | -0.1571 | No |
| 54 | Bcl10 | 11818 | -0.096 | -0.1569 | No |
| 55 | Ercc1 | 11880 | -0.099 | -0.1552 | No |
| 56 | Sod2 | 11918 | -0.100 | -0.1521 | No |
| 57 | Slc25a17 | 11955 | -0.102 | -0.1489 | No |
| 58 | Sod1 | 12072 | -0.108 | -0.1496 | No |
| 59 | Sema3c | 12110 | -0.109 | -0.1462 | No |
| 60 | Abcb9 | 12340 | -0.118 | -0.1523 | No |
| 61 | Cadm1 | 12644 | -0.131 | -0.1616 | No |
| 62 | Ide | 12714 | -0.135 | -0.1585 | No |
| 63 | Tspo | 12835 | -0.139 | -0.1578 | No |
| 64 | Cln8 | 13083 | -0.151 | -0.1632 | No |
| 65 | Fads1 | 13301 | -0.160 | -0.1666 | No |
| 66 | Cdk7 | 13840 | -0.183 | -0.1855 | No |
| 67 | Cln6 | 14283 | -0.204 | -0.1984 | No |
| 68 | Hsd11b2 | 14563 | -0.218 | -0.2021 | No |
| 69 | Mlycd | 14648 | -0.223 | -0.1955 | No |
| 70 | Esr2 | 15036 | -0.244 | -0.2036 | No |
| 71 | Cat | 15040 | -0.244 | -0.1916 | No |
| 72 | Fdps | 15598 | -0.274 | -0.2071 | No |
| 73 | Gstk1 | 15946 | -0.297 | -0.2104 | No |
| 74 | Cacna1b | 16261 | -0.319 | -0.2110 | No |
| 75 | Crat | 16416 | -0.329 | -0.2027 | No |
| 76 | Slc25a4 | 16839 | -0.358 | -0.2070 | No |
| 77 | Abcb4 | 17671 | -0.429 | -0.2290 | Yes |
| 78 | Pex11b | 17860 | -0.448 | -0.2166 | Yes |
| 79 | Idh1 | 18028 | -0.470 | -0.2020 | Yes |
| 80 | Nudt19 | 18061 | -0.475 | -0.1802 | Yes |
| 81 | Hras | 18354 | -0.524 | -0.1695 | Yes |
| 82 | Abcd1 | 18414 | -0.536 | -0.1460 | Yes |
| 83 | Hsd17b11 | 18447 | -0.543 | -0.1209 | Yes |
| 84 | Rdh11 | 18856 | -0.636 | -0.1106 | Yes |
| 85 | Slc35b2 | 19044 | -0.737 | -0.0839 | Yes |
| 86 | Pex11a | 19256 | -0.980 | -0.0464 | Yes |
| 87 | Hsd3b7 | 19259 | -0.990 | 0.0025 | Yes |