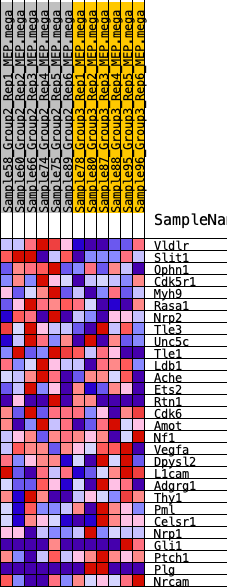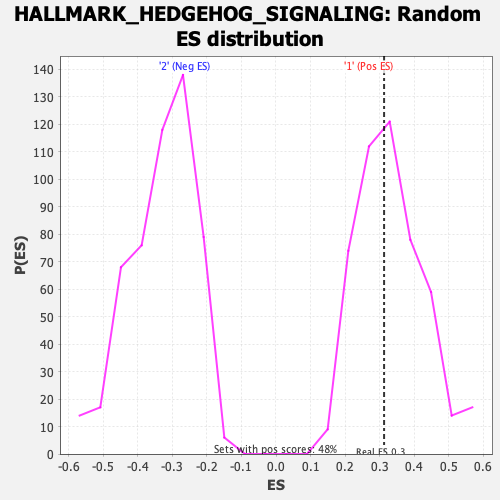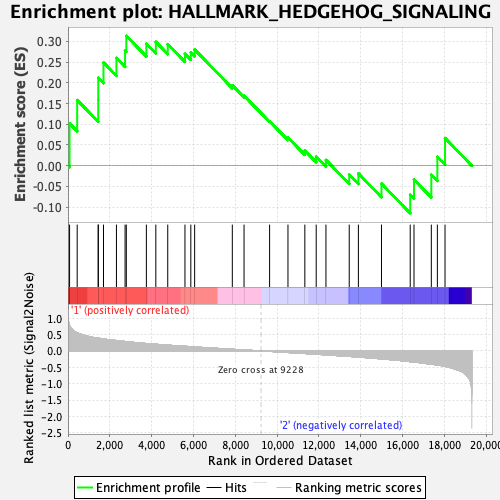
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_HEDGEHOG_SIGNALING |
| Enrichment Score (ES) | 0.3131447 |
| Normalized Enrichment Score (NES) | 0.94451094 |
| Nominal p-value | 0.5289256 |
| FDR q-value | 0.9420035 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Vldlr | 75 | 0.776 | 0.1017 | Yes |
| 2 | Slit1 | 439 | 0.554 | 0.1582 | Yes |
| 3 | Ophn1 | 1446 | 0.389 | 0.1590 | Yes |
| 4 | Cdk5r1 | 1447 | 0.389 | 0.2120 | Yes |
| 5 | Myh9 | 1697 | 0.365 | 0.2487 | Yes |
| 6 | Rasa1 | 2316 | 0.316 | 0.2597 | Yes |
| 7 | Nrp2 | 2729 | 0.288 | 0.2776 | Yes |
| 8 | Tle3 | 2788 | 0.284 | 0.3131 | Yes |
| 9 | Unc5c | 3747 | 0.226 | 0.2942 | No |
| 10 | Tle1 | 4199 | 0.205 | 0.2987 | No |
| 11 | Ldb1 | 4769 | 0.176 | 0.2931 | No |
| 12 | Ache | 5592 | 0.142 | 0.2699 | No |
| 13 | Ets2 | 5872 | 0.130 | 0.2731 | No |
| 14 | Rtn1 | 6055 | 0.121 | 0.2801 | No |
| 15 | Cdk6 | 7856 | 0.052 | 0.1938 | No |
| 16 | Amot | 8417 | 0.030 | 0.1688 | No |
| 17 | Nf1 | 9639 | -0.012 | 0.1071 | No |
| 18 | Vegfa | 10517 | -0.046 | 0.0679 | No |
| 19 | Dpysl2 | 11324 | -0.077 | 0.0365 | No |
| 20 | L1cam | 11867 | -0.099 | 0.0218 | No |
| 21 | Adgrg1 | 12333 | -0.117 | 0.0136 | No |
| 22 | Thy1 | 13446 | -0.165 | -0.0216 | No |
| 23 | Pml | 13884 | -0.186 | -0.0190 | No |
| 24 | Celsr1 | 14988 | -0.241 | -0.0434 | No |
| 25 | Nrp1 | 16362 | -0.326 | -0.0702 | No |
| 26 | Gli1 | 16543 | -0.337 | -0.0336 | No |
| 27 | Ptch1 | 17371 | -0.402 | -0.0218 | No |
| 28 | Plg | 17660 | -0.428 | 0.0214 | No |
| 29 | Nrcam | 18026 | -0.470 | 0.0664 | No |