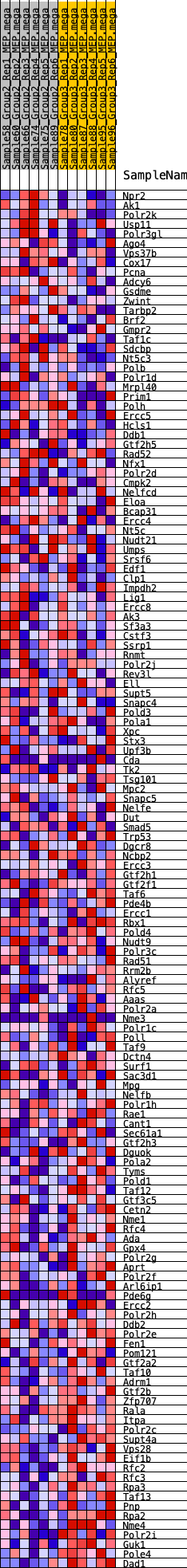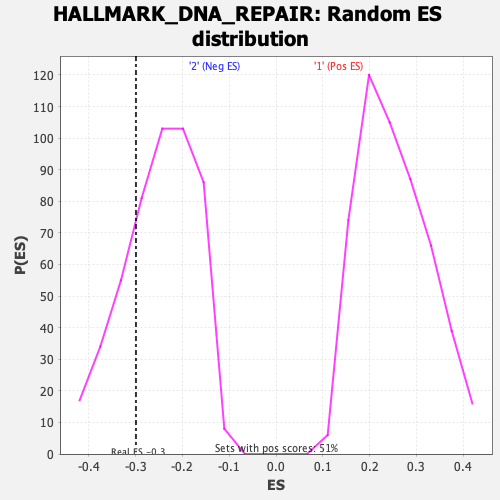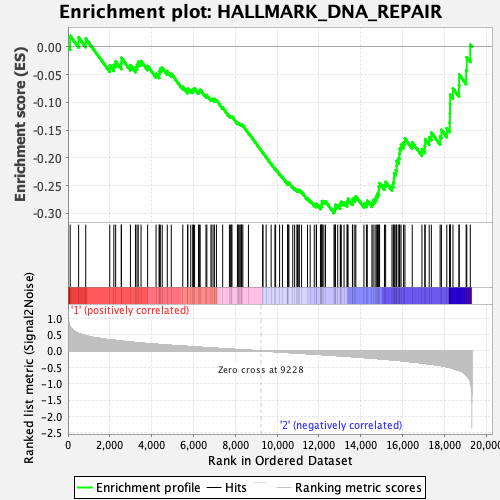
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_DNA_REPAIR |
| Enrichment Score (ES) | -0.29946047 |
| Normalized Enrichment Score (NES) | -1.1991463 |
| Nominal p-value | 0.24435319 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.933 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Npr2 | 105 | 0.739 | 0.0196 | No |
| 2 | Ak1 | 508 | 0.533 | 0.0167 | No |
| 3 | Polr2k | 847 | 0.465 | 0.0148 | No |
| 4 | Usp11 | 1996 | 0.343 | -0.0335 | No |
| 5 | Polr3gl | 2192 | 0.326 | -0.0326 | No |
| 6 | Ago4 | 2278 | 0.320 | -0.0262 | No |
| 7 | Vps37b | 2547 | 0.302 | -0.0299 | No |
| 8 | Cox17 | 2553 | 0.302 | -0.0200 | No |
| 9 | Pcna | 2984 | 0.271 | -0.0332 | No |
| 10 | Adcy6 | 3225 | 0.256 | -0.0371 | No |
| 11 | Gsdme | 3299 | 0.251 | -0.0324 | No |
| 12 | Zwint | 3358 | 0.247 | -0.0270 | No |
| 13 | Tarbp2 | 3492 | 0.240 | -0.0258 | No |
| 14 | Brf2 | 3807 | 0.222 | -0.0347 | No |
| 15 | Gmpr2 | 4211 | 0.204 | -0.0488 | No |
| 16 | Taf1c | 4358 | 0.197 | -0.0497 | No |
| 17 | Sdcbp | 4368 | 0.196 | -0.0436 | No |
| 18 | Nt5c3 | 4414 | 0.194 | -0.0393 | No |
| 19 | Polb | 4504 | 0.189 | -0.0376 | No |
| 20 | Polr1d | 4747 | 0.177 | -0.0442 | No |
| 21 | Mrpl40 | 4938 | 0.169 | -0.0484 | No |
| 22 | Prim1 | 5488 | 0.147 | -0.0721 | No |
| 23 | Polh | 5715 | 0.137 | -0.0792 | No |
| 24 | Ercc5 | 5732 | 0.137 | -0.0754 | No |
| 25 | Hcls1 | 5869 | 0.131 | -0.0781 | No |
| 26 | Ddb1 | 5962 | 0.126 | -0.0786 | No |
| 27 | Gtf2h5 | 6001 | 0.124 | -0.0764 | No |
| 28 | Rad52 | 6052 | 0.121 | -0.0749 | No |
| 29 | Nfx1 | 6238 | 0.113 | -0.0807 | No |
| 30 | Polr2d | 6280 | 0.112 | -0.0790 | No |
| 31 | Cmpk2 | 6319 | 0.111 | -0.0772 | No |
| 32 | Nelfcd | 6597 | 0.099 | -0.0883 | No |
| 33 | Eloa | 6638 | 0.098 | -0.0871 | No |
| 34 | Bcap31 | 6835 | 0.090 | -0.0943 | No |
| 35 | Ercc4 | 6885 | 0.088 | -0.0938 | No |
| 36 | Nt5c | 6983 | 0.084 | -0.0960 | No |
| 37 | Nudt21 | 6995 | 0.084 | -0.0938 | No |
| 38 | Umps | 7096 | 0.080 | -0.0963 | No |
| 39 | Srsf6 | 7395 | 0.068 | -0.1095 | No |
| 40 | Edf1 | 7717 | 0.057 | -0.1243 | No |
| 41 | Clp1 | 7778 | 0.054 | -0.1256 | No |
| 42 | Impdh2 | 7804 | 0.054 | -0.1251 | No |
| 43 | Lig1 | 7840 | 0.053 | -0.1252 | No |
| 44 | Ercc8 | 8101 | 0.042 | -0.1373 | No |
| 45 | Ak3 | 8132 | 0.041 | -0.1375 | No |
| 46 | Sf3a3 | 8168 | 0.039 | -0.1380 | No |
| 47 | Cstf3 | 8216 | 0.037 | -0.1392 | No |
| 48 | Ssrp1 | 8264 | 0.036 | -0.1404 | No |
| 49 | Rnmt | 8296 | 0.034 | -0.1409 | No |
| 50 | Polr2j | 8308 | 0.034 | -0.1403 | No |
| 51 | Rev3l | 8360 | 0.032 | -0.1419 | No |
| 52 | Ell | 8626 | 0.022 | -0.1550 | No |
| 53 | Supt5 | 9306 | -0.001 | -0.1904 | No |
| 54 | Snapc4 | 9323 | -0.001 | -0.1912 | No |
| 55 | Pold3 | 9473 | -0.006 | -0.1987 | No |
| 56 | Pola1 | 9713 | -0.014 | -0.2107 | No |
| 57 | Xpc | 9893 | -0.021 | -0.2193 | No |
| 58 | Stx3 | 9927 | -0.023 | -0.2203 | No |
| 59 | Upf3b | 10117 | -0.029 | -0.2292 | No |
| 60 | Cda | 10253 | -0.035 | -0.2350 | No |
| 61 | Tk2 | 10510 | -0.046 | -0.2468 | No |
| 62 | Tsg101 | 10512 | -0.046 | -0.2453 | No |
| 63 | Mpc2 | 10515 | -0.046 | -0.2439 | No |
| 64 | Snapc5 | 10563 | -0.048 | -0.2447 | No |
| 65 | Nelfe | 10747 | -0.054 | -0.2524 | No |
| 66 | Dut | 10836 | -0.058 | -0.2551 | No |
| 67 | Smad5 | 10944 | -0.061 | -0.2586 | No |
| 68 | Trp53 | 10976 | -0.063 | -0.2581 | No |
| 69 | Dgcr8 | 11038 | -0.065 | -0.2590 | No |
| 70 | Ncbp2 | 11058 | -0.066 | -0.2578 | No |
| 71 | Ercc3 | 11168 | -0.071 | -0.2611 | No |
| 72 | Gtf2h1 | 11451 | -0.080 | -0.2731 | No |
| 73 | Gtf2f1 | 11578 | -0.086 | -0.2767 | No |
| 74 | Taf6 | 11774 | -0.094 | -0.2837 | No |
| 75 | Pde4b | 11869 | -0.099 | -0.2853 | No |
| 76 | Ercc1 | 11880 | -0.099 | -0.2824 | No |
| 77 | Rbx1 | 12069 | -0.107 | -0.2886 | No |
| 78 | Pold4 | 12090 | -0.108 | -0.2860 | No |
| 79 | Nudt9 | 12138 | -0.110 | -0.2847 | No |
| 80 | Polr3c | 12141 | -0.110 | -0.2811 | No |
| 81 | Rad51 | 12148 | -0.110 | -0.2777 | No |
| 82 | Rrm2b | 12233 | -0.113 | -0.2782 | No |
| 83 | Alyref | 12310 | -0.116 | -0.2782 | No |
| 84 | Rfc5 | 12718 | -0.135 | -0.2949 | Yes |
| 85 | Aaas | 12758 | -0.136 | -0.2923 | Yes |
| 86 | Polr2a | 12780 | -0.137 | -0.2888 | Yes |
| 87 | Nme3 | 12787 | -0.137 | -0.2844 | Yes |
| 88 | Polr1c | 12895 | -0.142 | -0.2852 | Yes |
| 89 | Poll | 13018 | -0.148 | -0.2865 | Yes |
| 90 | Taf9 | 13027 | -0.149 | -0.2819 | Yes |
| 91 | Dctn4 | 13065 | -0.150 | -0.2787 | Yes |
| 92 | Surf1 | 13198 | -0.156 | -0.2803 | Yes |
| 93 | Sac3d1 | 13340 | -0.161 | -0.2822 | Yes |
| 94 | Mpg | 13365 | -0.162 | -0.2780 | Yes |
| 95 | Nelfb | 13389 | -0.163 | -0.2736 | Yes |
| 96 | Polr1h | 13600 | -0.173 | -0.2787 | Yes |
| 97 | Rae1 | 13616 | -0.174 | -0.2736 | Yes |
| 98 | Cant1 | 13714 | -0.178 | -0.2727 | Yes |
| 99 | Sec61a1 | 13763 | -0.180 | -0.2691 | Yes |
| 100 | Gtf2h3 | 14150 | -0.198 | -0.2825 | Yes |
| 101 | Dguok | 14268 | -0.204 | -0.2817 | Yes |
| 102 | Pola2 | 14309 | -0.206 | -0.2768 | Yes |
| 103 | Tyms | 14530 | -0.217 | -0.2809 | Yes |
| 104 | Pold1 | 14587 | -0.220 | -0.2764 | Yes |
| 105 | Taf12 | 14680 | -0.225 | -0.2736 | Yes |
| 106 | Gtf3c5 | 14740 | -0.228 | -0.2689 | Yes |
| 107 | Cetn2 | 14798 | -0.231 | -0.2641 | Yes |
| 108 | Nme1 | 14860 | -0.234 | -0.2593 | Yes |
| 109 | Rfc4 | 14862 | -0.234 | -0.2515 | Yes |
| 110 | Ada | 14898 | -0.237 | -0.2453 | Yes |
| 111 | Gpx4 | 15131 | -0.248 | -0.2490 | Yes |
| 112 | Polr2g | 15187 | -0.252 | -0.2433 | Yes |
| 113 | Aprt | 15491 | -0.269 | -0.2500 | Yes |
| 114 | Polr2f | 15554 | -0.271 | -0.2440 | Yes |
| 115 | Arl6ip1 | 15590 | -0.274 | -0.2366 | Yes |
| 116 | Pde6g | 15595 | -0.274 | -0.2275 | Yes |
| 117 | Ercc2 | 15675 | -0.278 | -0.2222 | Yes |
| 118 | Polr2h | 15713 | -0.281 | -0.2146 | Yes |
| 119 | Ddb2 | 15716 | -0.281 | -0.2052 | Yes |
| 120 | Polr2e | 15815 | -0.288 | -0.2006 | Yes |
| 121 | Fen1 | 15844 | -0.289 | -0.1922 | Yes |
| 122 | Pom121 | 15862 | -0.290 | -0.1833 | Yes |
| 123 | Gtf2a2 | 15923 | -0.296 | -0.1764 | Yes |
| 124 | Taf10 | 16045 | -0.304 | -0.1724 | Yes |
| 125 | Adrm1 | 16109 | -0.309 | -0.1652 | Yes |
| 126 | Gtf2b | 16459 | -0.332 | -0.1722 | Yes |
| 127 | Zfp707 | 16923 | -0.365 | -0.1840 | Yes |
| 128 | Rala | 17062 | -0.378 | -0.1784 | Yes |
| 129 | Itpa | 17075 | -0.378 | -0.1662 | Yes |
| 130 | Polr2c | 17270 | -0.396 | -0.1629 | Yes |
| 131 | Supt4a | 17372 | -0.402 | -0.1545 | Yes |
| 132 | Vps28 | 17792 | -0.440 | -0.1615 | Yes |
| 133 | Eif1b | 17854 | -0.447 | -0.1495 | Yes |
| 134 | Rfc2 | 18111 | -0.483 | -0.1465 | Yes |
| 135 | Rfc3 | 18250 | -0.504 | -0.1366 | Yes |
| 136 | Rpa3 | 18260 | -0.505 | -0.1200 | Yes |
| 137 | Taf13 | 18266 | -0.506 | -0.1031 | Yes |
| 138 | Pnp | 18273 | -0.506 | -0.0862 | Yes |
| 139 | Rpa2 | 18406 | -0.535 | -0.0750 | Yes |
| 140 | Nme4 | 18694 | -0.594 | -0.0698 | Yes |
| 141 | Polr2i | 18703 | -0.598 | -0.0500 | Yes |
| 142 | Guk1 | 19036 | -0.731 | -0.0425 | Yes |
| 143 | Pole4 | 19066 | -0.748 | -0.0187 | Yes |
| 144 | Dad1 | 19234 | -0.921 | 0.0038 | Yes |