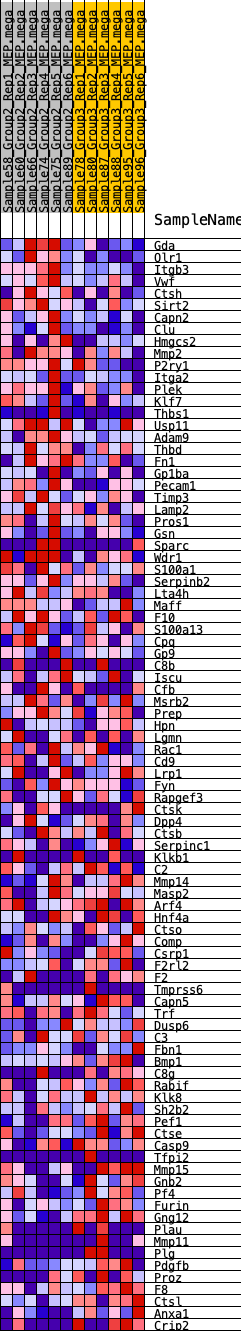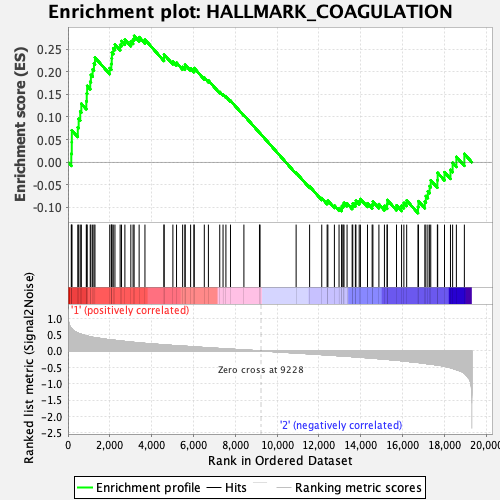
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.27937365 |
| Normalized Enrichment Score (NES) | 0.95370394 |
| Nominal p-value | 0.5728347 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gda | 157 | 0.689 | 0.0189 | Yes |
| 2 | Olr1 | 180 | 0.675 | 0.0442 | Yes |
| 3 | Itgb3 | 183 | 0.674 | 0.0706 | Yes |
| 4 | Vwf | 468 | 0.543 | 0.0772 | Yes |
| 5 | Ctsh | 510 | 0.532 | 0.0959 | Yes |
| 6 | Sirt2 | 585 | 0.515 | 0.1123 | Yes |
| 7 | Capn2 | 638 | 0.506 | 0.1295 | Yes |
| 8 | Clu | 875 | 0.459 | 0.1353 | Yes |
| 9 | Hmgcs2 | 896 | 0.455 | 0.1521 | Yes |
| 10 | Mmp2 | 916 | 0.452 | 0.1689 | Yes |
| 11 | P2ry1 | 1063 | 0.429 | 0.1781 | Yes |
| 12 | Itga2 | 1095 | 0.424 | 0.1932 | Yes |
| 13 | Plek | 1178 | 0.415 | 0.2052 | Yes |
| 14 | Klf7 | 1241 | 0.409 | 0.2181 | Yes |
| 15 | Thbs1 | 1289 | 0.403 | 0.2315 | Yes |
| 16 | Usp11 | 1996 | 0.343 | 0.2082 | Yes |
| 17 | Adam9 | 2070 | 0.336 | 0.2176 | Yes |
| 18 | Thbd | 2090 | 0.333 | 0.2297 | Yes |
| 19 | Fn1 | 2100 | 0.332 | 0.2423 | Yes |
| 20 | Gp1ba | 2163 | 0.330 | 0.2520 | Yes |
| 21 | Pecam1 | 2244 | 0.323 | 0.2605 | Yes |
| 22 | Timp3 | 2493 | 0.305 | 0.2595 | Yes |
| 23 | Lamp2 | 2559 | 0.301 | 0.2680 | Yes |
| 24 | Pros1 | 2720 | 0.289 | 0.2710 | Yes |
| 25 | Gsn | 3004 | 0.270 | 0.2669 | Yes |
| 26 | Sparc | 3111 | 0.264 | 0.2717 | Yes |
| 27 | Wdr1 | 3161 | 0.260 | 0.2794 | Yes |
| 28 | S100a1 | 3401 | 0.245 | 0.2766 | No |
| 29 | Serpinb2 | 3679 | 0.230 | 0.2712 | No |
| 30 | Lta4h | 4590 | 0.184 | 0.2311 | No |
| 31 | Maff | 4592 | 0.184 | 0.2382 | No |
| 32 | F10 | 5015 | 0.166 | 0.2228 | No |
| 33 | S100a13 | 5189 | 0.159 | 0.2200 | No |
| 34 | Cpq | 5481 | 0.147 | 0.2107 | No |
| 35 | Gp9 | 5594 | 0.142 | 0.2104 | No |
| 36 | C8b | 5598 | 0.142 | 0.2158 | No |
| 37 | Iscu | 5862 | 0.131 | 0.2073 | No |
| 38 | Cfb | 6023 | 0.123 | 0.2038 | No |
| 39 | Msrb2 | 6032 | 0.123 | 0.2082 | No |
| 40 | Prep | 6517 | 0.103 | 0.1871 | No |
| 41 | Hpn | 6713 | 0.095 | 0.1806 | No |
| 42 | Lgmn | 7255 | 0.073 | 0.1554 | No |
| 43 | Rac1 | 7413 | 0.068 | 0.1499 | No |
| 44 | Cd9 | 7550 | 0.062 | 0.1452 | No |
| 45 | Lrp1 | 7769 | 0.055 | 0.1360 | No |
| 46 | Fyn | 8410 | 0.030 | 0.1039 | No |
| 47 | Rapgef3 | 9155 | 0.003 | 0.0653 | No |
| 48 | Ctsk | 9182 | 0.002 | 0.0641 | No |
| 49 | Dpp4 | 10907 | -0.060 | -0.0233 | No |
| 50 | Ctsb | 11552 | -0.085 | -0.0535 | No |
| 51 | Serpinc1 | 12132 | -0.110 | -0.0793 | No |
| 52 | Klkb1 | 12390 | -0.120 | -0.0880 | No |
| 53 | C2 | 12425 | -0.121 | -0.0850 | No |
| 54 | Mmp14 | 12737 | -0.136 | -0.0958 | No |
| 55 | Masp2 | 12959 | -0.145 | -0.1016 | No |
| 56 | Arf4 | 13082 | -0.151 | -0.1021 | No |
| 57 | Hnf4a | 13098 | -0.152 | -0.0969 | No |
| 58 | Ctso | 13155 | -0.154 | -0.0937 | No |
| 59 | Comp | 13196 | -0.156 | -0.0897 | No |
| 60 | Csrp1 | 13343 | -0.161 | -0.0910 | No |
| 61 | F2rl2 | 13586 | -0.172 | -0.0968 | No |
| 62 | F2 | 13615 | -0.174 | -0.0914 | No |
| 63 | Tmprss6 | 13740 | -0.179 | -0.0909 | No |
| 64 | Capn5 | 13766 | -0.180 | -0.0851 | No |
| 65 | Trf | 13923 | -0.188 | -0.0858 | No |
| 66 | Dusp6 | 13981 | -0.190 | -0.0813 | No |
| 67 | C3 | 14319 | -0.206 | -0.0908 | No |
| 68 | Fbn1 | 14542 | -0.217 | -0.0938 | No |
| 69 | Bmp1 | 14578 | -0.219 | -0.0870 | No |
| 70 | C8g | 14865 | -0.234 | -0.0927 | No |
| 71 | Rabif | 15129 | -0.248 | -0.0966 | No |
| 72 | Klk8 | 15256 | -0.257 | -0.0931 | No |
| 73 | Sh2b2 | 15266 | -0.257 | -0.0835 | No |
| 74 | Pef1 | 15707 | -0.280 | -0.0954 | No |
| 75 | Ctse | 15948 | -0.297 | -0.0962 | No |
| 76 | Casp9 | 16059 | -0.306 | -0.0899 | No |
| 77 | Tfpi2 | 16193 | -0.314 | -0.0845 | No |
| 78 | Mmp15 | 16734 | -0.350 | -0.0988 | No |
| 79 | Gnb2 | 16756 | -0.352 | -0.0861 | No |
| 80 | Pf4 | 17061 | -0.377 | -0.0871 | No |
| 81 | Furin | 17111 | -0.382 | -0.0747 | No |
| 82 | Gng12 | 17207 | -0.390 | -0.0643 | No |
| 83 | Plau | 17290 | -0.398 | -0.0529 | No |
| 84 | Mmp11 | 17344 | -0.400 | -0.0400 | No |
| 85 | Plg | 17660 | -0.428 | -0.0396 | No |
| 86 | Pdgfb | 17675 | -0.430 | -0.0234 | No |
| 87 | Proz | 18001 | -0.466 | -0.0220 | No |
| 88 | F8 | 18287 | -0.511 | -0.0168 | No |
| 89 | Ctsl | 18390 | -0.531 | -0.0013 | No |
| 90 | Anxa1 | 18569 | -0.569 | 0.0118 | No |
| 91 | Crip2 | 18951 | -0.675 | 0.0185 | No |