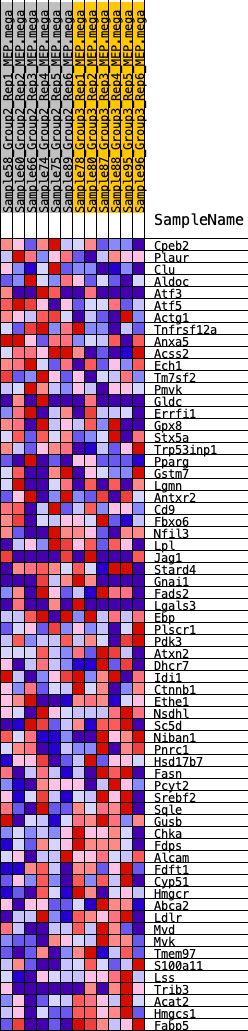
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
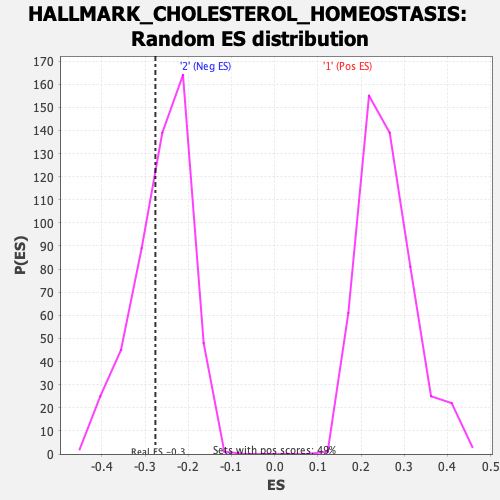
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | -0.27616537 |
| Normalized Enrichment Score (NES) | -1.0638523 |
| Nominal p-value | 0.34697855 |
| FDR q-value | 0.8452324 |
| FWER p-Value | 0.977 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Cpeb2 | 619 | 0.509 | -0.0017 | No |
| 2 | Plaur | 827 | 0.469 | 0.0156 | No |
| 3 | Clu | 875 | 0.459 | 0.0406 | No |
| 4 | Aldoc | 993 | 0.440 | 0.0608 | No |
| 5 | Atf3 | 1187 | 0.414 | 0.0756 | No |
| 6 | Atf5 | 1345 | 0.398 | 0.0913 | No |
| 7 | Actg1 | 1530 | 0.382 | 0.1046 | No |
| 8 | Tnfrsf12a | 1599 | 0.376 | 0.1235 | No |
| 9 | Anxa5 | 1660 | 0.368 | 0.1424 | No |
| 10 | Acss2 | 2332 | 0.316 | 0.1265 | No |
| 11 | Ech1 | 2987 | 0.271 | 0.1087 | No |
| 12 | Tm7sf2 | 3275 | 0.253 | 0.1089 | No |
| 13 | Pmvk | 3334 | 0.249 | 0.1208 | No |
| 14 | Gldc | 3800 | 0.223 | 0.1099 | No |
| 15 | Errfi1 | 4182 | 0.205 | 0.1024 | No |
| 16 | Gpx8 | 4502 | 0.189 | 0.0971 | No |
| 17 | Stx5a | 4897 | 0.171 | 0.0869 | No |
| 18 | Trp53inp1 | 4963 | 0.168 | 0.0936 | No |
| 19 | Pparg | 5366 | 0.152 | 0.0818 | No |
| 20 | Gstm7 | 6989 | 0.084 | 0.0025 | No |
| 21 | Lgmn | 7255 | 0.073 | -0.0069 | No |
| 22 | Antxr2 | 7329 | 0.071 | -0.0065 | No |
| 23 | Cd9 | 7550 | 0.062 | -0.0142 | No |
| 24 | Fbxo6 | 7949 | 0.048 | -0.0319 | No |
| 25 | Nfil3 | 8164 | 0.040 | -0.0407 | No |
| 26 | Lpl | 8774 | 0.017 | -0.0713 | No |
| 27 | Jag1 | 9416 | -0.005 | -0.1043 | No |
| 28 | Stard4 | 9497 | -0.007 | -0.1081 | No |
| 29 | Gnai1 | 9552 | -0.010 | -0.1103 | No |
| 30 | Fads2 | 9697 | -0.014 | -0.1169 | No |
| 31 | Lgals3 | 9702 | -0.014 | -0.1163 | No |
| 32 | Ebp | 9791 | -0.017 | -0.1198 | No |
| 33 | Plscr1 | 9824 | -0.018 | -0.1204 | No |
| 34 | Pdk3 | 10403 | -0.041 | -0.1480 | No |
| 35 | Atxn2 | 11228 | -0.073 | -0.1865 | No |
| 36 | Dhcr7 | 11530 | -0.084 | -0.1971 | No |
| 37 | Idi1 | 11730 | -0.092 | -0.2019 | No |
| 38 | Ctnnb1 | 12258 | -0.114 | -0.2224 | No |
| 39 | Ethe1 | 12532 | -0.126 | -0.2291 | No |
| 40 | Nsdhl | 12811 | -0.138 | -0.2352 | No |
| 41 | Sc5d | 13412 | -0.164 | -0.2566 | No |
| 42 | Niban1 | 13529 | -0.169 | -0.2525 | No |
| 43 | Pnrc1 | 13623 | -0.174 | -0.2469 | No |
| 44 | Hsd17b7 | 13896 | -0.186 | -0.2499 | No |
| 45 | Fasn | 14311 | -0.206 | -0.2591 | No |
| 46 | Pcyt2 | 14640 | -0.223 | -0.2629 | Yes |
| 47 | Srebf2 | 14690 | -0.225 | -0.2519 | Yes |
| 48 | Sqle | 14971 | -0.241 | -0.2521 | Yes |
| 49 | Gusb | 15054 | -0.245 | -0.2417 | Yes |
| 50 | Chka | 15456 | -0.267 | -0.2466 | Yes |
| 51 | Fdps | 15598 | -0.274 | -0.2375 | Yes |
| 52 | Alcam | 15665 | -0.278 | -0.2243 | Yes |
| 53 | Fdft1 | 15826 | -0.288 | -0.2154 | Yes |
| 54 | Cyp51 | 16143 | -0.311 | -0.2132 | Yes |
| 55 | Hmgcr | 16418 | -0.329 | -0.2078 | Yes |
| 56 | Abca2 | 16596 | -0.341 | -0.1966 | Yes |
| 57 | Ldlr | 16762 | -0.352 | -0.1841 | Yes |
| 58 | Mvd | 16959 | -0.368 | -0.1723 | Yes |
| 59 | Mvk | 17566 | -0.421 | -0.1786 | Yes |
| 60 | Tmem97 | 18005 | -0.467 | -0.1735 | Yes |
| 61 | S100a11 | 18091 | -0.479 | -0.1492 | Yes |
| 62 | Lss | 18391 | -0.531 | -0.1330 | Yes |
| 63 | Trib3 | 18652 | -0.585 | -0.1115 | Yes |
| 64 | Acat2 | 19112 | -0.786 | -0.0884 | Yes |
| 65 | Hmgcs1 | 19137 | -0.806 | -0.0414 | Yes |
| 66 | Fabp5 | 19176 | -0.839 | 0.0068 | Yes |