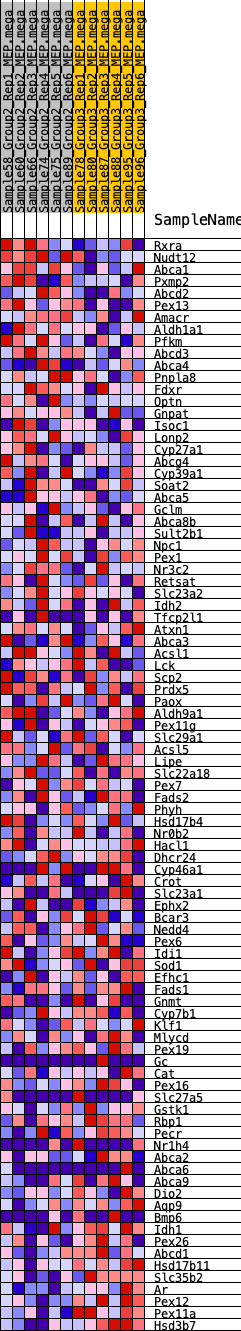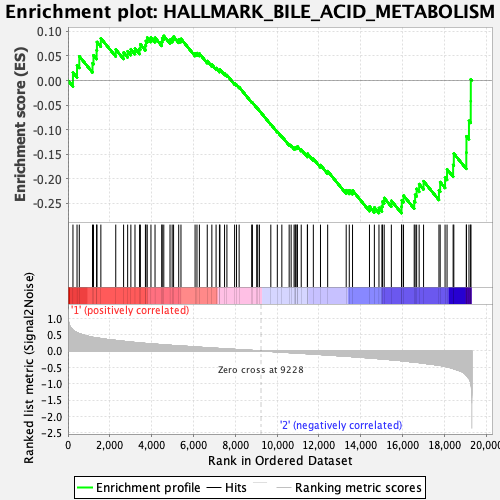
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group2_versus_Group3.MEP.mega_Pheno.cls #Group2_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group2_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.26967856 |
| Normalized Enrichment Score (NES) | -1.1717255 |
| Nominal p-value | 0.18074656 |
| FDR q-value | 0.9125132 |
| FWER p-Value | 0.945 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rxra | 237 | 0.630 | 0.0161 | No |
| 2 | Nudt12 | 432 | 0.557 | 0.0311 | No |
| 3 | Abca1 | 540 | 0.526 | 0.0492 | No |
| 4 | Pxmp2 | 1172 | 0.416 | 0.0351 | No |
| 5 | Abcd2 | 1218 | 0.411 | 0.0513 | No |
| 6 | Pex13 | 1368 | 0.396 | 0.0614 | No |
| 7 | Amacr | 1384 | 0.394 | 0.0784 | No |
| 8 | Aldh1a1 | 1571 | 0.379 | 0.0858 | No |
| 9 | Pfkm | 2283 | 0.320 | 0.0632 | No |
| 10 | Abcd3 | 2658 | 0.294 | 0.0570 | No |
| 11 | Abca4 | 2852 | 0.279 | 0.0595 | No |
| 12 | Pnpla8 | 3006 | 0.270 | 0.0637 | No |
| 13 | Fdxr | 3203 | 0.257 | 0.0651 | No |
| 14 | Optn | 3421 | 0.244 | 0.0648 | No |
| 15 | Gnpat | 3461 | 0.241 | 0.0736 | No |
| 16 | Isoc1 | 3701 | 0.229 | 0.0715 | No |
| 17 | Lonp2 | 3725 | 0.227 | 0.0806 | No |
| 18 | Cyp27a1 | 3788 | 0.224 | 0.0874 | No |
| 19 | Abcg4 | 3966 | 0.214 | 0.0878 | No |
| 20 | Cyp39a1 | 4161 | 0.207 | 0.0871 | No |
| 21 | Soat2 | 4483 | 0.190 | 0.0789 | No |
| 22 | Abca5 | 4503 | 0.189 | 0.0864 | No |
| 23 | Gclm | 4580 | 0.185 | 0.0908 | No |
| 24 | Abca8b | 4878 | 0.172 | 0.0831 | No |
| 25 | Sult2b1 | 4980 | 0.167 | 0.0854 | No |
| 26 | Npc1 | 5048 | 0.164 | 0.0893 | No |
| 27 | Pex1 | 5293 | 0.155 | 0.0836 | No |
| 28 | Nr3c2 | 5401 | 0.150 | 0.0848 | No |
| 29 | Retsat | 6076 | 0.120 | 0.0551 | No |
| 30 | Slc23a2 | 6166 | 0.117 | 0.0557 | No |
| 31 | Idh2 | 6282 | 0.112 | 0.0548 | No |
| 32 | Tfcp2l1 | 6658 | 0.097 | 0.0397 | No |
| 33 | Atxn1 | 6878 | 0.089 | 0.0323 | No |
| 34 | Abca3 | 7080 | 0.081 | 0.0254 | No |
| 35 | Acsl1 | 7241 | 0.074 | 0.0204 | No |
| 36 | Lck | 7264 | 0.073 | 0.0226 | No |
| 37 | Scp2 | 7485 | 0.065 | 0.0141 | No |
| 38 | Prdx5 | 7598 | 0.060 | 0.0110 | No |
| 39 | Paox | 7966 | 0.047 | -0.0060 | No |
| 40 | Aldh9a1 | 8058 | 0.043 | -0.0088 | No |
| 41 | Pex11g | 8182 | 0.039 | -0.0134 | No |
| 42 | Slc29a1 | 8783 | 0.017 | -0.0439 | No |
| 43 | Acsl5 | 8817 | 0.015 | -0.0449 | No |
| 44 | Lipe | 9023 | 0.008 | -0.0552 | No |
| 45 | Slc22a18 | 9071 | 0.006 | -0.0574 | No |
| 46 | Pex7 | 9152 | 0.003 | -0.0614 | No |
| 47 | Fads2 | 9697 | -0.014 | -0.0891 | No |
| 48 | Phyh | 10008 | -0.026 | -0.1041 | No |
| 49 | Hsd17b4 | 10224 | -0.034 | -0.1137 | No |
| 50 | Nr0b2 | 10574 | -0.048 | -0.1297 | No |
| 51 | Hacl1 | 10669 | -0.051 | -0.1323 | No |
| 52 | Dhcr24 | 10809 | -0.056 | -0.1370 | No |
| 53 | Cyp46a1 | 10862 | -0.059 | -0.1371 | No |
| 54 | Crot | 10893 | -0.060 | -0.1359 | No |
| 55 | Slc23a1 | 10954 | -0.062 | -0.1363 | No |
| 56 | Ephx2 | 10968 | -0.062 | -0.1341 | No |
| 57 | Bcar3 | 11151 | -0.070 | -0.1404 | No |
| 58 | Nedd4 | 11443 | -0.080 | -0.1520 | No |
| 59 | Pex6 | 11452 | -0.081 | -0.1487 | No |
| 60 | Idi1 | 11730 | -0.092 | -0.1590 | No |
| 61 | Sod1 | 12072 | -0.108 | -0.1719 | No |
| 62 | Efhc1 | 12415 | -0.121 | -0.1842 | No |
| 63 | Fads1 | 13301 | -0.160 | -0.2231 | No |
| 64 | Gnmt | 13448 | -0.165 | -0.2232 | No |
| 65 | Cyp7b1 | 13604 | -0.173 | -0.2235 | No |
| 66 | Klf1 | 14414 | -0.212 | -0.2561 | No |
| 67 | Mlycd | 14648 | -0.223 | -0.2581 | No |
| 68 | Pex19 | 14868 | -0.234 | -0.2590 | Yes |
| 69 | Gc | 15010 | -0.243 | -0.2554 | Yes |
| 70 | Cat | 15040 | -0.244 | -0.2459 | Yes |
| 71 | Pex16 | 15119 | -0.247 | -0.2388 | Yes |
| 72 | Slc27a5 | 15460 | -0.268 | -0.2444 | Yes |
| 73 | Gstk1 | 15946 | -0.297 | -0.2563 | Yes |
| 74 | Rbp1 | 15960 | -0.299 | -0.2435 | Yes |
| 75 | Pecr | 16042 | -0.304 | -0.2340 | Yes |
| 76 | Nr1h4 | 16553 | -0.338 | -0.2453 | Yes |
| 77 | Abca2 | 16596 | -0.341 | -0.2321 | Yes |
| 78 | Abca6 | 16669 | -0.345 | -0.2203 | Yes |
| 79 | Abca9 | 16789 | -0.354 | -0.2106 | Yes |
| 80 | Dio2 | 17000 | -0.372 | -0.2047 | Yes |
| 81 | Aqp9 | 17736 | -0.436 | -0.2233 | Yes |
| 82 | Bmp6 | 17794 | -0.440 | -0.2065 | Yes |
| 83 | Idh1 | 18028 | -0.470 | -0.1974 | Yes |
| 84 | Pex26 | 18123 | -0.484 | -0.1805 | Yes |
| 85 | Abcd1 | 18414 | -0.536 | -0.1714 | Yes |
| 86 | Hsd17b11 | 18447 | -0.543 | -0.1486 | Yes |
| 87 | Slc35b2 | 19044 | -0.737 | -0.1464 | Yes |
| 88 | Ar | 19051 | -0.740 | -0.1134 | Yes |
| 89 | Pex12 | 19171 | -0.837 | -0.0818 | Yes |
| 90 | Pex11a | 19256 | -0.980 | -0.0420 | Yes |
| 91 | Hsd3b7 | 19259 | -0.990 | 0.0025 | Yes |