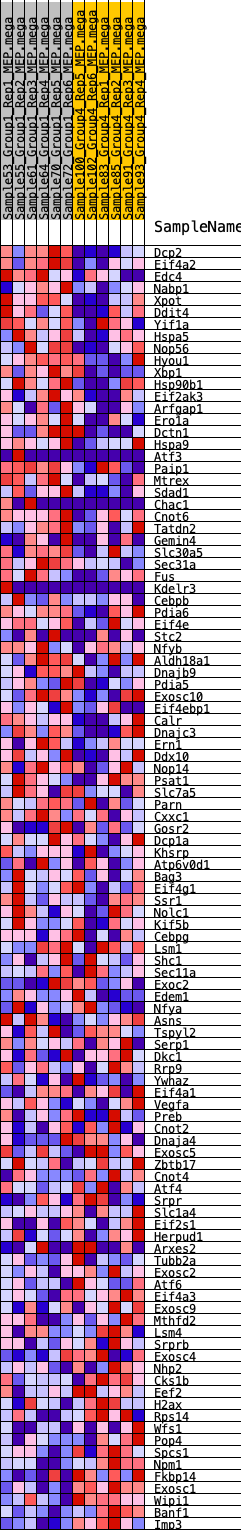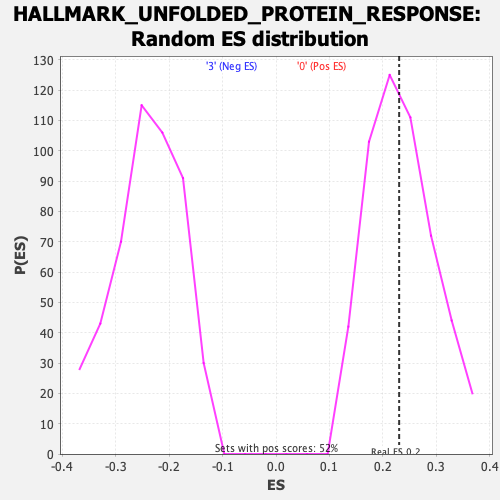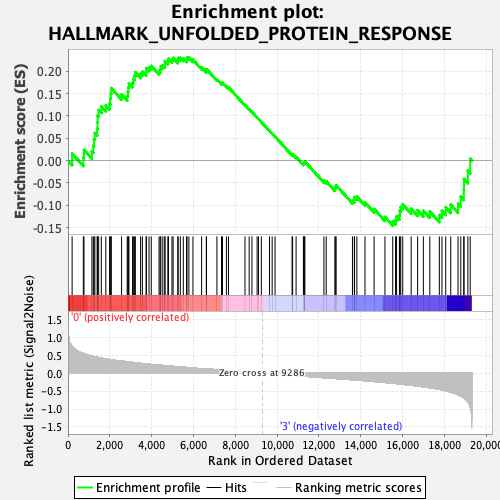
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.23071857 |
| Normalized Enrichment Score (NES) | 0.9870235 |
| Nominal p-value | 0.49516442 |
| FDR q-value | 0.80082023 |
| FWER p-Value | 0.988 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Dcp2 | 197 | 0.744 | 0.0155 | Yes |
| 2 | Eif4a2 | 733 | 0.546 | 0.0066 | Yes |
| 3 | Edc4 | 764 | 0.538 | 0.0237 | Yes |
| 4 | Nabp1 | 1143 | 0.478 | 0.0206 | Yes |
| 5 | Xpot | 1210 | 0.467 | 0.0333 | Yes |
| 6 | Ddit4 | 1243 | 0.462 | 0.0477 | Yes |
| 7 | Yif1a | 1284 | 0.458 | 0.0615 | Yes |
| 8 | Hspa5 | 1393 | 0.442 | 0.0712 | Yes |
| 9 | Nop56 | 1412 | 0.439 | 0.0855 | Yes |
| 10 | Hyou1 | 1415 | 0.439 | 0.1006 | Yes |
| 11 | Xbp1 | 1467 | 0.433 | 0.1129 | Yes |
| 12 | Hsp90b1 | 1594 | 0.420 | 0.1209 | Yes |
| 13 | Eif2ak3 | 1808 | 0.394 | 0.1235 | Yes |
| 14 | Arfgap1 | 1989 | 0.379 | 0.1273 | Yes |
| 15 | Ero1a | 2019 | 0.374 | 0.1387 | Yes |
| 16 | Dctn1 | 2046 | 0.372 | 0.1503 | Yes |
| 17 | Hspa9 | 2069 | 0.369 | 0.1619 | Yes |
| 18 | Atf3 | 2561 | 0.334 | 0.1479 | Yes |
| 19 | Paip1 | 2831 | 0.313 | 0.1448 | Yes |
| 20 | Mtrex | 2866 | 0.311 | 0.1538 | Yes |
| 21 | Sdad1 | 2882 | 0.309 | 0.1637 | Yes |
| 22 | Chac1 | 2913 | 0.307 | 0.1728 | Yes |
| 23 | Cnot6 | 3085 | 0.297 | 0.1742 | Yes |
| 24 | Tatdn2 | 3124 | 0.295 | 0.1824 | Yes |
| 25 | Gemin4 | 3184 | 0.291 | 0.1894 | Yes |
| 26 | Slc30a5 | 3220 | 0.288 | 0.1976 | Yes |
| 27 | Sec31a | 3471 | 0.272 | 0.1940 | Yes |
| 28 | Fus | 3556 | 0.268 | 0.1989 | Yes |
| 29 | Kdelr3 | 3735 | 0.255 | 0.1985 | Yes |
| 30 | Cebpb | 3748 | 0.254 | 0.2067 | Yes |
| 31 | Pdia6 | 3874 | 0.247 | 0.2087 | Yes |
| 32 | Eif4e | 3981 | 0.240 | 0.2115 | Yes |
| 33 | Stc2 | 4354 | 0.224 | 0.1999 | Yes |
| 34 | Nfyb | 4406 | 0.221 | 0.2049 | Yes |
| 35 | Aldh18a1 | 4439 | 0.219 | 0.2108 | Yes |
| 36 | Dnajb9 | 4533 | 0.214 | 0.2134 | Yes |
| 37 | Pdia5 | 4631 | 0.207 | 0.2155 | Yes |
| 38 | Exosc10 | 4635 | 0.207 | 0.2225 | Yes |
| 39 | Eif4ebp1 | 4775 | 0.201 | 0.2223 | Yes |
| 40 | Calr | 4802 | 0.199 | 0.2278 | Yes |
| 41 | Dnajc3 | 4971 | 0.190 | 0.2257 | Yes |
| 42 | Ern1 | 5024 | 0.189 | 0.2295 | Yes |
| 43 | Ddx10 | 5250 | 0.179 | 0.2240 | Yes |
| 44 | Nop14 | 5265 | 0.179 | 0.2295 | Yes |
| 45 | Psat1 | 5370 | 0.173 | 0.2301 | Yes |
| 46 | Slc7a5 | 5513 | 0.165 | 0.2284 | Yes |
| 47 | Parn | 5669 | 0.157 | 0.2258 | Yes |
| 48 | Cxxc1 | 5689 | 0.156 | 0.2302 | Yes |
| 49 | Gosr2 | 5780 | 0.151 | 0.2307 | Yes |
| 50 | Dcp1a | 5975 | 0.141 | 0.2255 | No |
| 51 | Khsrp | 6388 | 0.123 | 0.2083 | No |
| 52 | Atp6v0d1 | 6614 | 0.119 | 0.2007 | No |
| 53 | Bag3 | 6620 | 0.119 | 0.2046 | No |
| 54 | Eif4g1 | 7120 | 0.095 | 0.1819 | No |
| 55 | Ssr1 | 7341 | 0.084 | 0.1733 | No |
| 56 | Nolc1 | 7386 | 0.082 | 0.1739 | No |
| 57 | Kif5b | 7575 | 0.074 | 0.1667 | No |
| 58 | Cebpg | 7674 | 0.069 | 0.1640 | No |
| 59 | Lsm1 | 8462 | 0.035 | 0.1242 | No |
| 60 | Shc1 | 8662 | 0.027 | 0.1148 | No |
| 61 | Sec11a | 8795 | 0.022 | 0.1086 | No |
| 62 | Exoc2 | 9042 | 0.011 | 0.0962 | No |
| 63 | Edem1 | 9106 | 0.008 | 0.0932 | No |
| 64 | Nfya | 9243 | 0.002 | 0.0862 | No |
| 65 | Asns | 9634 | -0.009 | 0.0662 | No |
| 66 | Tspyl2 | 9761 | -0.014 | 0.0601 | No |
| 67 | Serp1 | 9900 | -0.021 | 0.0537 | No |
| 68 | Dkc1 | 10714 | -0.058 | 0.0133 | No |
| 69 | Rrp9 | 10740 | -0.059 | 0.0141 | No |
| 70 | Ywhaz | 10905 | -0.067 | 0.0079 | No |
| 71 | Eif4a1 | 11255 | -0.082 | -0.0074 | No |
| 72 | Vegfa | 11285 | -0.084 | -0.0060 | No |
| 73 | Preb | 11293 | -0.084 | -0.0035 | No |
| 74 | Cnot2 | 11325 | -0.086 | -0.0021 | No |
| 75 | Dnaja4 | 12236 | -0.125 | -0.0452 | No |
| 76 | Exosc5 | 12345 | -0.130 | -0.0463 | No |
| 77 | Zbtb17 | 12764 | -0.147 | -0.0629 | No |
| 78 | Cnot4 | 12779 | -0.148 | -0.0585 | No |
| 79 | Atf4 | 12820 | -0.150 | -0.0554 | No |
| 80 | Srpr | 13600 | -0.183 | -0.0896 | No |
| 81 | Slc1a4 | 13691 | -0.187 | -0.0878 | No |
| 82 | Eif2s1 | 13701 | -0.187 | -0.0818 | No |
| 83 | Herpud1 | 13813 | -0.192 | -0.0809 | No |
| 84 | Arxes2 | 14199 | -0.209 | -0.0937 | No |
| 85 | Tubb2a | 14635 | -0.233 | -0.1083 | No |
| 86 | Exosc2 | 15154 | -0.259 | -0.1263 | No |
| 87 | Atf6 | 15526 | -0.280 | -0.1359 | No |
| 88 | Eif4a3 | 15662 | -0.288 | -0.1329 | No |
| 89 | Exosc9 | 15709 | -0.291 | -0.1253 | No |
| 90 | Mthfd2 | 15846 | -0.299 | -0.1220 | No |
| 91 | Lsm4 | 15867 | -0.301 | -0.1126 | No |
| 92 | Srprb | 15909 | -0.304 | -0.1042 | No |
| 93 | Exosc4 | 16004 | -0.311 | -0.0983 | No |
| 94 | Nhp2 | 16407 | -0.337 | -0.1076 | No |
| 95 | Cks1b | 16716 | -0.359 | -0.1112 | No |
| 96 | Eef2 | 16990 | -0.380 | -0.1122 | No |
| 97 | H2ax | 17299 | -0.408 | -0.1141 | No |
| 98 | Rps14 | 17754 | -0.448 | -0.1222 | No |
| 99 | Wfs1 | 17879 | -0.464 | -0.1126 | No |
| 100 | Pop4 | 18059 | -0.491 | -0.1049 | No |
| 101 | Spcs1 | 18300 | -0.530 | -0.0990 | No |
| 102 | Npm1 | 18649 | -0.600 | -0.0964 | No |
| 103 | Fkbp14 | 18780 | -0.638 | -0.0810 | No |
| 104 | Exosc1 | 18924 | -0.683 | -0.0648 | No |
| 105 | Wipi1 | 18933 | -0.686 | -0.0414 | No |
| 106 | Banf1 | 19118 | -0.808 | -0.0230 | No |
| 107 | Imp3 | 19230 | -0.946 | 0.0040 | No |