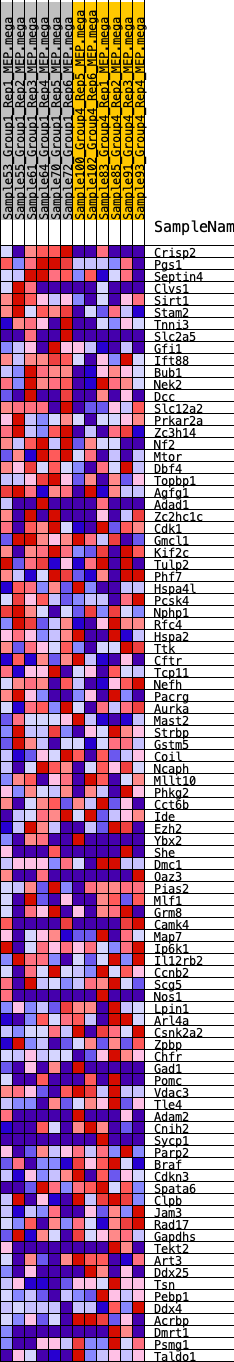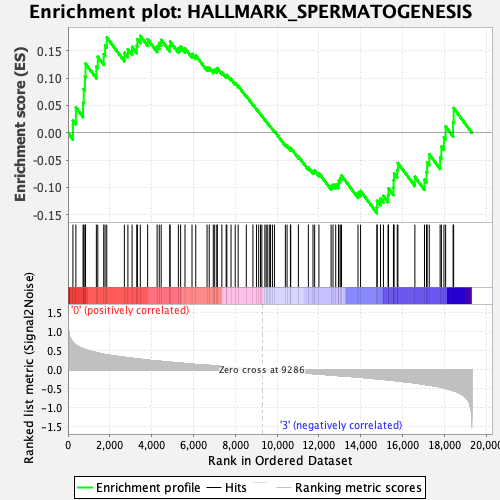
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.17701232 |
| Normalized Enrichment Score (NES) | 0.7390295 |
| Nominal p-value | 0.8822355 |
| FDR q-value | 0.9406292 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Crisp2 | 235 | 0.718 | 0.0225 | Yes |
| 2 | Pgs1 | 376 | 0.644 | 0.0464 | Yes |
| 3 | Septin4 | 720 | 0.549 | 0.0552 | Yes |
| 4 | Clvs1 | 753 | 0.542 | 0.0798 | Yes |
| 5 | Sirt1 | 810 | 0.529 | 0.1025 | Yes |
| 6 | Stam2 | 839 | 0.525 | 0.1265 | Yes |
| 7 | Tnni3 | 1356 | 0.446 | 0.1212 | Yes |
| 8 | Slc2a5 | 1424 | 0.438 | 0.1389 | Yes |
| 9 | Gfi1 | 1708 | 0.404 | 0.1437 | Yes |
| 10 | Ift88 | 1773 | 0.397 | 0.1596 | Yes |
| 11 | Bub1 | 1854 | 0.390 | 0.1743 | Yes |
| 12 | Nek2 | 2695 | 0.326 | 0.1464 | Yes |
| 13 | Dcc | 2864 | 0.311 | 0.1527 | Yes |
| 14 | Slc12a2 | 3064 | 0.299 | 0.1569 | Yes |
| 15 | Prkar2a | 3287 | 0.283 | 0.1590 | Yes |
| 16 | Zc3h14 | 3318 | 0.280 | 0.1710 | Yes |
| 17 | Nf2 | 3457 | 0.273 | 0.1770 | Yes |
| 18 | Mtor | 3807 | 0.251 | 0.1710 | No |
| 19 | Dbf4 | 4261 | 0.228 | 0.1584 | No |
| 20 | Topbp1 | 4363 | 0.223 | 0.1640 | No |
| 21 | Agfg1 | 4455 | 0.218 | 0.1698 | No |
| 22 | Adad1 | 4866 | 0.196 | 0.1580 | No |
| 23 | Zc2hc1c | 4881 | 0.196 | 0.1667 | No |
| 24 | Cdk1 | 5277 | 0.178 | 0.1548 | No |
| 25 | Gmcl1 | 5385 | 0.172 | 0.1575 | No |
| 26 | Kif2c | 5594 | 0.161 | 0.1545 | No |
| 27 | Tulp2 | 5929 | 0.143 | 0.1441 | No |
| 28 | Phf7 | 6108 | 0.134 | 0.1413 | No |
| 29 | Hspa4l | 6648 | 0.117 | 0.1190 | No |
| 30 | Pcsk4 | 6752 | 0.112 | 0.1190 | No |
| 31 | Nphp1 | 6946 | 0.103 | 0.1140 | No |
| 32 | Rfc4 | 7014 | 0.099 | 0.1153 | No |
| 33 | Hspa2 | 7117 | 0.095 | 0.1146 | No |
| 34 | Ttk | 7139 | 0.094 | 0.1180 | No |
| 35 | Cftr | 7356 | 0.083 | 0.1108 | No |
| 36 | Tcp11 | 7562 | 0.074 | 0.1038 | No |
| 37 | Nefh | 7595 | 0.073 | 0.1056 | No |
| 38 | Pacrg | 7795 | 0.064 | 0.0984 | No |
| 39 | Aurka | 7994 | 0.054 | 0.0907 | No |
| 40 | Mast2 | 8135 | 0.049 | 0.0857 | No |
| 41 | Strbp | 8525 | 0.033 | 0.0671 | No |
| 42 | Gstm5 | 8841 | 0.020 | 0.0516 | No |
| 43 | Coil | 9011 | 0.012 | 0.0434 | No |
| 44 | Ncaph | 9116 | 0.007 | 0.0384 | No |
| 45 | Mllt10 | 9205 | 0.004 | 0.0340 | No |
| 46 | Phkg2 | 9269 | 0.001 | 0.0307 | No |
| 47 | Cct6b | 9414 | -0.001 | 0.0233 | No |
| 48 | Ide | 9477 | -0.003 | 0.0202 | No |
| 49 | Ezh2 | 9552 | -0.006 | 0.0166 | No |
| 50 | Ybx2 | 9645 | -0.009 | 0.0123 | No |
| 51 | She | 9680 | -0.011 | 0.0111 | No |
| 52 | Dmc1 | 9774 | -0.015 | 0.0069 | No |
| 53 | Oaz3 | 9870 | -0.020 | 0.0030 | No |
| 54 | Pias2 | 10392 | -0.044 | -0.0220 | No |
| 55 | Mlf1 | 10474 | -0.047 | -0.0240 | No |
| 56 | Grm8 | 10634 | -0.054 | -0.0296 | No |
| 57 | Camk4 | 10649 | -0.055 | -0.0277 | No |
| 58 | Map7 | 11015 | -0.072 | -0.0432 | No |
| 59 | Ip6k1 | 11492 | -0.094 | -0.0634 | No |
| 60 | Il12rb2 | 11714 | -0.103 | -0.0699 | No |
| 61 | Ccnb2 | 11793 | -0.107 | -0.0688 | No |
| 62 | Scg5 | 11997 | -0.116 | -0.0738 | No |
| 63 | Nos1 | 12579 | -0.140 | -0.0972 | No |
| 64 | Lpin1 | 12670 | -0.144 | -0.0950 | No |
| 65 | Arl4a | 12800 | -0.150 | -0.0944 | No |
| 66 | Csnk2a2 | 12932 | -0.155 | -0.0937 | No |
| 67 | Zpbp | 12949 | -0.156 | -0.0870 | No |
| 68 | Chfr | 13018 | -0.158 | -0.0829 | No |
| 69 | Gad1 | 13077 | -0.161 | -0.0781 | No |
| 70 | Pomc | 13866 | -0.195 | -0.1097 | No |
| 71 | Vdac3 | 13988 | -0.201 | -0.1062 | No |
| 72 | Tle4 | 14770 | -0.240 | -0.1353 | No |
| 73 | Adam2 | 14780 | -0.241 | -0.1241 | No |
| 74 | Cnih2 | 14941 | -0.248 | -0.1204 | No |
| 75 | Sycp1 | 15081 | -0.256 | -0.1152 | No |
| 76 | Parp2 | 15303 | -0.269 | -0.1137 | No |
| 77 | Braf | 15325 | -0.270 | -0.1017 | No |
| 78 | Cdkn3 | 15560 | -0.282 | -0.1002 | No |
| 79 | Spata6 | 15567 | -0.283 | -0.0869 | No |
| 80 | Clpb | 15591 | -0.284 | -0.0743 | No |
| 81 | Jam3 | 15742 | -0.293 | -0.0679 | No |
| 82 | Rad17 | 15773 | -0.294 | -0.0552 | No |
| 83 | Gapdhs | 16585 | -0.349 | -0.0805 | No |
| 84 | Tekt2 | 17044 | -0.386 | -0.0857 | No |
| 85 | Art3 | 17140 | -0.396 | -0.0714 | No |
| 86 | Ddx25 | 17173 | -0.397 | -0.0539 | No |
| 87 | Tsn | 17273 | -0.406 | -0.0393 | No |
| 88 | Pebp1 | 17792 | -0.453 | -0.0444 | No |
| 89 | Ddx4 | 17853 | -0.461 | -0.0252 | No |
| 90 | Acrbp | 17974 | -0.480 | -0.0081 | No |
| 91 | Dmrt1 | 18055 | -0.490 | 0.0114 | No |
| 92 | Psmg1 | 18412 | -0.550 | 0.0195 | No |
| 93 | Taldo1 | 18439 | -0.557 | 0.0452 | No |