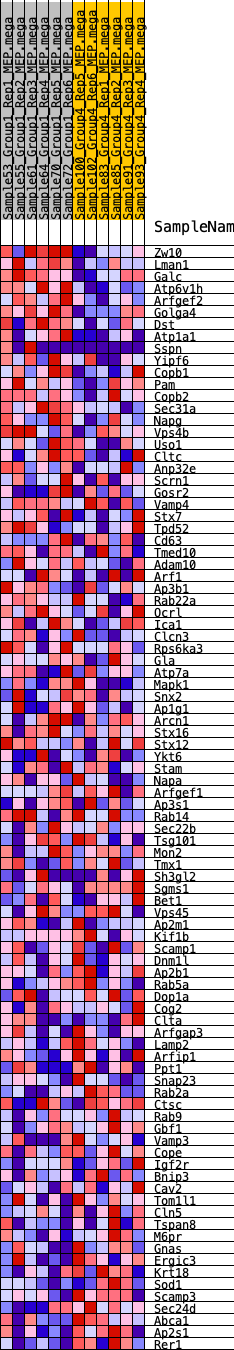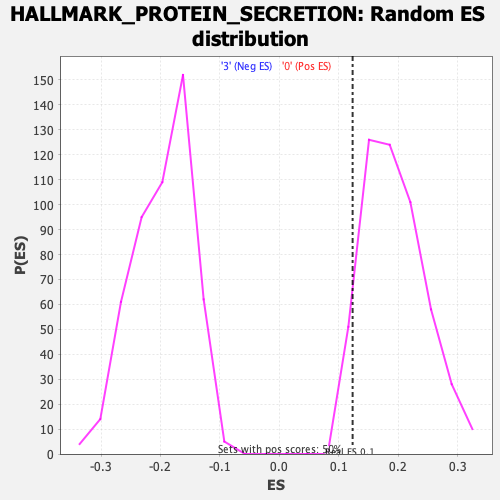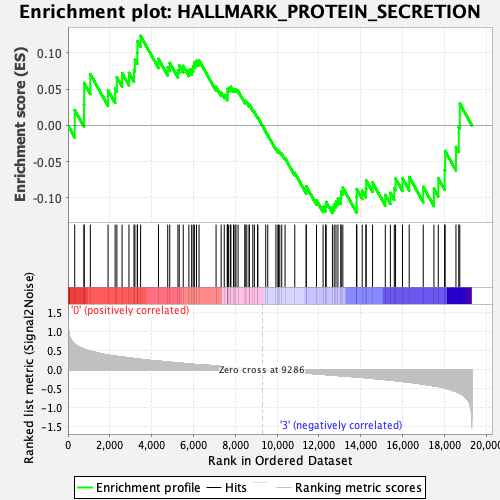
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.12329854 |
| Normalized Enrichment Score (NES) | 0.6395245 |
| Nominal p-value | 0.939759 |
| FDR q-value | 0.93560064 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Zw10 | 321 | 0.670 | 0.0211 | Yes |
| 2 | Lman1 | 762 | 0.539 | 0.0285 | Yes |
| 3 | Galc | 774 | 0.537 | 0.0582 | Yes |
| 4 | Atp6v1h | 1068 | 0.489 | 0.0706 | Yes |
| 5 | Arfgef2 | 1915 | 0.384 | 0.0482 | Yes |
| 6 | Golga4 | 2252 | 0.357 | 0.0508 | Yes |
| 7 | Dst | 2336 | 0.351 | 0.0662 | Yes |
| 8 | Atp1a1 | 2585 | 0.332 | 0.0721 | Yes |
| 9 | Sspn | 2915 | 0.307 | 0.0723 | Yes |
| 10 | Yipf6 | 3155 | 0.293 | 0.0763 | Yes |
| 11 | Copb1 | 3199 | 0.290 | 0.0904 | Yes |
| 12 | Pam | 3313 | 0.280 | 0.1003 | Yes |
| 13 | Copb2 | 3320 | 0.280 | 0.1158 | Yes |
| 14 | Sec31a | 3471 | 0.272 | 0.1233 | Yes |
| 15 | Napg | 4326 | 0.226 | 0.0916 | No |
| 16 | Vps4b | 4768 | 0.201 | 0.0800 | No |
| 17 | Uso1 | 4865 | 0.196 | 0.0860 | No |
| 18 | Cltc | 5255 | 0.179 | 0.0759 | No |
| 19 | Anp32e | 5318 | 0.175 | 0.0825 | No |
| 20 | Scrn1 | 5510 | 0.165 | 0.0819 | No |
| 21 | Gosr2 | 5780 | 0.151 | 0.0764 | No |
| 22 | Vamp4 | 5912 | 0.144 | 0.0777 | No |
| 23 | Stx7 | 5986 | 0.140 | 0.0818 | No |
| 24 | Tpd52 | 6043 | 0.137 | 0.0866 | No |
| 25 | Cd63 | 6143 | 0.132 | 0.0889 | No |
| 26 | Tmed10 | 6265 | 0.127 | 0.0898 | No |
| 27 | Adam10 | 7079 | 0.097 | 0.0530 | No |
| 28 | Arf1 | 7322 | 0.085 | 0.0451 | No |
| 29 | Ap3b1 | 7471 | 0.079 | 0.0419 | No |
| 30 | Rab22a | 7613 | 0.072 | 0.0386 | No |
| 31 | Ocrl | 7625 | 0.071 | 0.0420 | No |
| 32 | Ica1 | 7628 | 0.071 | 0.0459 | No |
| 33 | Clcn3 | 7633 | 0.071 | 0.0497 | No |
| 34 | Rps6ka3 | 7680 | 0.069 | 0.0513 | No |
| 35 | Gla | 7763 | 0.065 | 0.0507 | No |
| 36 | Atp7a | 7788 | 0.064 | 0.0530 | No |
| 37 | Mapk1 | 7915 | 0.058 | 0.0497 | No |
| 38 | Snx2 | 7982 | 0.054 | 0.0493 | No |
| 39 | Ap1g1 | 8030 | 0.053 | 0.0498 | No |
| 40 | Arcn1 | 8132 | 0.049 | 0.0473 | No |
| 41 | Stx16 | 8459 | 0.035 | 0.0323 | No |
| 42 | Stx12 | 8468 | 0.035 | 0.0339 | No |
| 43 | Ykt6 | 8549 | 0.032 | 0.0315 | No |
| 44 | Stam | 8660 | 0.027 | 0.0273 | No |
| 45 | Napa | 8666 | 0.027 | 0.0286 | No |
| 46 | Arfgef1 | 8835 | 0.020 | 0.0210 | No |
| 47 | Ap3s1 | 8912 | 0.017 | 0.0180 | No |
| 48 | Rab14 | 9066 | 0.010 | 0.0106 | No |
| 49 | Sec22b | 9075 | 0.010 | 0.0107 | No |
| 50 | Tsg101 | 9453 | -0.002 | -0.0088 | No |
| 51 | Mon2 | 9551 | -0.005 | -0.0135 | No |
| 52 | Tmx1 | 9941 | -0.023 | -0.0325 | No |
| 53 | Sh3gl2 | 10036 | -0.029 | -0.0357 | No |
| 54 | Sgms1 | 10061 | -0.030 | -0.0353 | No |
| 55 | Bet1 | 10125 | -0.033 | -0.0367 | No |
| 56 | Vps45 | 10219 | -0.038 | -0.0395 | No |
| 57 | Ap2m1 | 10377 | -0.043 | -0.0452 | No |
| 58 | Kif1b | 10842 | -0.065 | -0.0657 | No |
| 59 | Scamp1 | 11389 | -0.089 | -0.0891 | No |
| 60 | Dnm1l | 11391 | -0.089 | -0.0842 | No |
| 61 | Ap2b1 | 11882 | -0.111 | -0.1034 | No |
| 62 | Rab5a | 12196 | -0.124 | -0.1127 | No |
| 63 | Dop1a | 12316 | -0.129 | -0.1116 | No |
| 64 | Cog2 | 12344 | -0.130 | -0.1057 | No |
| 65 | Clta | 12643 | -0.142 | -0.1132 | No |
| 66 | Arfgap3 | 12726 | -0.146 | -0.1092 | No |
| 67 | Lamp2 | 12812 | -0.150 | -0.1052 | No |
| 68 | Arfip1 | 12906 | -0.154 | -0.1013 | No |
| 69 | Ppt1 | 13047 | -0.160 | -0.0996 | No |
| 70 | Snap23 | 13058 | -0.160 | -0.0911 | No |
| 71 | Rab2a | 13141 | -0.164 | -0.0862 | No |
| 72 | Ctsc | 13798 | -0.192 | -0.1095 | No |
| 73 | Rab9 | 13799 | -0.192 | -0.0987 | No |
| 74 | Gbf1 | 13804 | -0.192 | -0.0881 | No |
| 75 | Vamp3 | 14067 | -0.206 | -0.0901 | No |
| 76 | Cope | 14244 | -0.211 | -0.0874 | No |
| 77 | Igf2r | 14256 | -0.212 | -0.0761 | No |
| 78 | Bnip3 | 14561 | -0.229 | -0.0790 | No |
| 79 | Cav2 | 15170 | -0.260 | -0.0960 | No |
| 80 | Tom1l1 | 15414 | -0.275 | -0.0931 | No |
| 81 | Cln5 | 15601 | -0.285 | -0.0868 | No |
| 82 | Tspan8 | 15661 | -0.288 | -0.0736 | No |
| 83 | M6pr | 15992 | -0.310 | -0.0733 | No |
| 84 | Gnas | 16313 | -0.331 | -0.0713 | No |
| 85 | Ergic3 | 16985 | -0.380 | -0.0848 | No |
| 86 | Krt18 | 17496 | -0.425 | -0.0874 | No |
| 87 | Sod1 | 17704 | -0.442 | -0.0733 | No |
| 88 | Scamp3 | 18006 | -0.484 | -0.0617 | No |
| 89 | Sec24d | 18031 | -0.486 | -0.0355 | No |
| 90 | Abca1 | 18549 | -0.575 | -0.0300 | No |
| 91 | Ap2s1 | 18683 | -0.608 | -0.0027 | No |
| 92 | Rer1 | 18732 | -0.623 | 0.0299 | No |