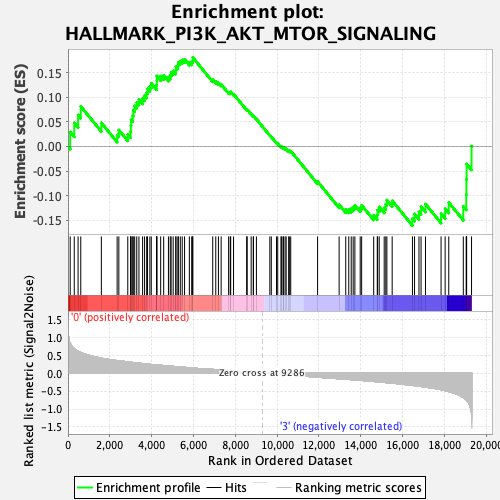
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PI3K_AKT_MTOR_SIGNALING |
| Enrichment Score (ES) | 0.18098338 |
| Normalized Enrichment Score (NES) | 0.7729442 |
| Nominal p-value | 0.8079208 |
| FDR q-value | 0.94314253 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Prkcb | 105 | 0.832 | 0.0293 | Yes |
| 2 | Trib3 | 299 | 0.684 | 0.0479 | Yes |
| 3 | Ralb | 482 | 0.611 | 0.0640 | Yes |
| 4 | Il4 | 610 | 0.577 | 0.0816 | Yes |
| 5 | Hsp90b1 | 1594 | 0.420 | 0.0480 | Yes |
| 6 | Tsc2 | 2349 | 0.350 | 0.0234 | Yes |
| 7 | Plcb1 | 2431 | 0.343 | 0.0335 | Yes |
| 8 | Pikfyve | 2858 | 0.311 | 0.0244 | Yes |
| 9 | Mknk2 | 2989 | 0.305 | 0.0304 | Yes |
| 10 | Sla | 3006 | 0.304 | 0.0422 | Yes |
| 11 | Traf2 | 3016 | 0.302 | 0.0544 | Yes |
| 12 | Mapk8 | 3095 | 0.296 | 0.0628 | Yes |
| 13 | Mknk1 | 3120 | 0.295 | 0.0739 | Yes |
| 14 | Cdk2 | 3189 | 0.290 | 0.0825 | Yes |
| 15 | Prkar2a | 3287 | 0.283 | 0.0892 | Yes |
| 16 | Fgf17 | 3392 | 0.276 | 0.0954 | Yes |
| 17 | Nfkbib | 3567 | 0.267 | 0.0975 | Yes |
| 18 | Smad2 | 3661 | 0.259 | 0.1035 | Yes |
| 19 | Sfn | 3762 | 0.254 | 0.1089 | Yes |
| 20 | Actr2 | 3803 | 0.251 | 0.1173 | Yes |
| 21 | Actr3 | 3914 | 0.244 | 0.1218 | Yes |
| 22 | Eif4e | 3981 | 0.240 | 0.1284 | Yes |
| 23 | Nck1 | 4235 | 0.229 | 0.1248 | Yes |
| 24 | Mapkap1 | 4245 | 0.228 | 0.1339 | Yes |
| 25 | Prkaa2 | 4248 | 0.228 | 0.1434 | Yes |
| 26 | Ptpn11 | 4431 | 0.219 | 0.1431 | Yes |
| 27 | Ube2n | 4575 | 0.211 | 0.1444 | Yes |
| 28 | Calr | 4802 | 0.199 | 0.1410 | Yes |
| 29 | Vav3 | 4888 | 0.195 | 0.1447 | Yes |
| 30 | Plcg1 | 4930 | 0.193 | 0.1507 | Yes |
| 31 | Nod1 | 5028 | 0.189 | 0.1536 | Yes |
| 32 | Ppp2r1b | 5153 | 0.185 | 0.1548 | Yes |
| 33 | Grb2 | 5162 | 0.184 | 0.1621 | Yes |
| 34 | Cltc | 5255 | 0.179 | 0.1648 | Yes |
| 35 | Cdk1 | 5277 | 0.178 | 0.1712 | Yes |
| 36 | Akt1 | 5369 | 0.173 | 0.1737 | Yes |
| 37 | Cab39 | 5461 | 0.168 | 0.1760 | Yes |
| 38 | Rptor | 5573 | 0.162 | 0.1770 | Yes |
| 39 | Ripk1 | 5802 | 0.150 | 0.1714 | Yes |
| 40 | Ywhab | 5914 | 0.144 | 0.1716 | Yes |
| 41 | Raf1 | 5962 | 0.142 | 0.1751 | Yes |
| 42 | Map2k3 | 5964 | 0.142 | 0.1810 | Yes |
| 43 | Irak4 | 6920 | 0.104 | 0.1356 | No |
| 44 | Atf1 | 7065 | 0.097 | 0.1322 | No |
| 45 | Ube2d3 | 7192 | 0.091 | 0.1295 | No |
| 46 | Arf1 | 7322 | 0.085 | 0.1263 | No |
| 47 | Rps6ka3 | 7680 | 0.069 | 0.1106 | No |
| 48 | Pik3r3 | 7757 | 0.066 | 0.1094 | No |
| 49 | Pten | 7779 | 0.064 | 0.1110 | No |
| 50 | Mapk1 | 7915 | 0.058 | 0.1064 | No |
| 51 | Pla2g12a | 8542 | 0.032 | 0.0751 | No |
| 52 | Rit1 | 8567 | 0.032 | 0.0752 | No |
| 53 | Cdkn1a | 8757 | 0.023 | 0.0663 | No |
| 54 | Acaca | 8860 | 0.019 | 0.0618 | No |
| 55 | Rps6ka1 | 8865 | 0.019 | 0.0624 | No |
| 56 | Map3k7 | 9008 | 0.013 | 0.0555 | No |
| 57 | Mapk9 | 9651 | -0.010 | 0.0225 | No |
| 58 | Gsk3b | 9720 | -0.013 | 0.0195 | No |
| 59 | Tiam1 | 9970 | -0.025 | 0.0076 | No |
| 60 | Lck | 10027 | -0.029 | 0.0059 | No |
| 61 | Pdk1 | 10182 | -0.036 | -0.0007 | No |
| 62 | Cab39l | 10215 | -0.037 | -0.0008 | No |
| 63 | Cxcr4 | 10285 | -0.040 | -0.0027 | No |
| 64 | Myd88 | 10299 | -0.041 | -0.0016 | No |
| 65 | Ap2m1 | 10377 | -0.043 | -0.0039 | No |
| 66 | Prkag1 | 10431 | -0.045 | -0.0047 | No |
| 67 | Cdkn1b | 10549 | -0.051 | -0.0087 | No |
| 68 | Tbk1 | 10599 | -0.053 | -0.0090 | No |
| 69 | Camk4 | 10649 | -0.055 | -0.0093 | No |
| 70 | Dusp3 | 11933 | -0.113 | -0.0713 | No |
| 71 | Itpr2 | 12963 | -0.157 | -0.1184 | No |
| 72 | Map2k6 | 13279 | -0.170 | -0.1276 | No |
| 73 | E2f1 | 13424 | -0.175 | -0.1278 | No |
| 74 | Gna14 | 13541 | -0.181 | -0.1263 | No |
| 75 | Pin1 | 13634 | -0.184 | -0.1234 | No |
| 76 | Ddit3 | 13716 | -0.188 | -0.1197 | No |
| 77 | Pitx2 | 13971 | -0.200 | -0.1245 | No |
| 78 | Adcy2 | 14040 | -0.204 | -0.1196 | No |
| 79 | Dapp1 | 14616 | -0.231 | -0.1398 | No |
| 80 | Mapk10 | 14787 | -0.241 | -0.1386 | No |
| 81 | Pak4 | 14803 | -0.242 | -0.1292 | No |
| 82 | Arhgdia | 14880 | -0.246 | -0.1229 | No |
| 83 | Rac1 | 15118 | -0.257 | -0.1244 | No |
| 84 | Ecsit | 15194 | -0.262 | -0.1174 | No |
| 85 | Stat2 | 15246 | -0.266 | -0.1089 | No |
| 86 | Csnk2b | 15499 | -0.278 | -0.1104 | No |
| 87 | Cdk4 | 16469 | -0.341 | -0.1466 | No |
| 88 | Tnfrsf1a | 16569 | -0.348 | -0.1371 | No |
| 89 | Akt1s1 | 16781 | -0.364 | -0.1329 | No |
| 90 | Grk2 | 16880 | -0.372 | -0.1224 | No |
| 91 | Pfn1 | 17092 | -0.391 | -0.1171 | No |
| 92 | Arpc3 | 17837 | -0.459 | -0.1366 | No |
| 93 | Ppp1ca | 18032 | -0.486 | -0.1263 | No |
| 94 | Il2rg | 18205 | -0.512 | -0.1139 | No |
| 95 | Slc2a1 | 18900 | -0.673 | -0.1218 | No |
| 96 | Them4 | 19041 | -0.747 | -0.0979 | No |
| 97 | Sqstm1 | 19048 | -0.750 | -0.0668 | No |
| 98 | Cfl1 | 19060 | -0.760 | -0.0356 | No |
| 99 | Hras | 19294 | -1.156 | 0.0007 | No |

