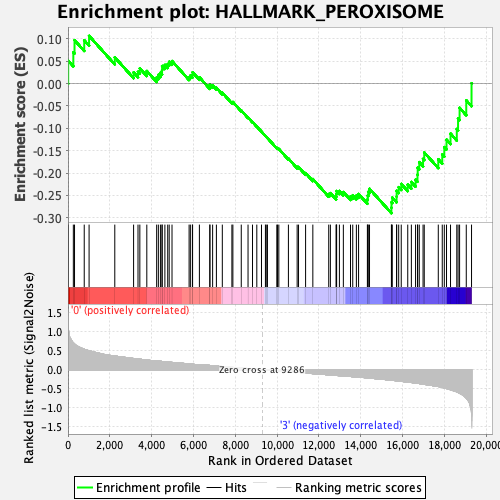
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.28967115 |
| Normalized Enrichment Score (NES) | -1.3387418 |
| Nominal p-value | 0.057026476 |
| FDR q-value | 0.57387763 |
| FWER p-Value | 0.695 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Abcb1a | 9 | 1.167 | 0.0513 | No |
| 2 | Fdps | 255 | 0.705 | 0.0699 | No |
| 3 | Gnpat | 312 | 0.674 | 0.0969 | No |
| 4 | Hsd11b2 | 776 | 0.536 | 0.0967 | No |
| 5 | Cacna1b | 1008 | 0.496 | 0.1066 | No |
| 6 | Sema3c | 2237 | 0.358 | 0.0587 | No |
| 7 | Msh2 | 3136 | 0.294 | 0.0250 | No |
| 8 | Slc23a2 | 3347 | 0.278 | 0.0264 | No |
| 9 | Cln8 | 3435 | 0.274 | 0.0341 | No |
| 10 | Pex6 | 3768 | 0.253 | 0.0281 | No |
| 11 | Acsl1 | 4239 | 0.229 | 0.0138 | No |
| 12 | Slc35b2 | 4320 | 0.226 | 0.0196 | No |
| 13 | Itgb1bp1 | 4419 | 0.220 | 0.0243 | No |
| 14 | Pex5 | 4496 | 0.216 | 0.0299 | No |
| 15 | Smarcc1 | 4501 | 0.216 | 0.0393 | No |
| 16 | Slc25a19 | 4632 | 0.207 | 0.0417 | No |
| 17 | Vps4b | 4768 | 0.201 | 0.0437 | No |
| 18 | Esr2 | 4838 | 0.198 | 0.0488 | No |
| 19 | Ercc3 | 4973 | 0.190 | 0.0503 | No |
| 20 | Acox1 | 5790 | 0.150 | 0.0145 | No |
| 21 | Aldh9a1 | 5850 | 0.147 | 0.0180 | No |
| 22 | Pex2 | 5947 | 0.143 | 0.0193 | No |
| 23 | Abcb4 | 5957 | 0.142 | 0.0252 | No |
| 24 | Cadm1 | 6282 | 0.127 | 0.0140 | No |
| 25 | Crat | 6781 | 0.111 | -0.0070 | No |
| 26 | Fads1 | 6785 | 0.111 | -0.0023 | No |
| 27 | Aldh1a1 | 6915 | 0.104 | -0.0044 | No |
| 28 | Dhrs3 | 7095 | 0.096 | -0.0094 | No |
| 29 | Top2a | 7378 | 0.082 | -0.0204 | No |
| 30 | Idh2 | 7836 | 0.062 | -0.0415 | No |
| 31 | Siah1a | 7883 | 0.059 | -0.0412 | No |
| 32 | Dhcr24 | 8284 | 0.041 | -0.0602 | No |
| 33 | Cnbp | 8608 | 0.029 | -0.0757 | No |
| 34 | Hmgcl | 8822 | 0.020 | -0.0859 | No |
| 35 | Acsl5 | 9027 | 0.012 | -0.0960 | No |
| 36 | Mvp | 9251 | 0.002 | -0.1075 | No |
| 37 | Fis1 | 9439 | -0.002 | -0.1172 | No |
| 38 | Ide | 9477 | -0.003 | -0.1190 | No |
| 39 | Acsl4 | 9536 | -0.005 | -0.1218 | No |
| 40 | Abcd3 | 9984 | -0.026 | -0.1439 | No |
| 41 | Hsd3b7 | 10018 | -0.028 | -0.1443 | No |
| 42 | Acot8 | 10080 | -0.031 | -0.1461 | No |
| 43 | Elovl5 | 10538 | -0.050 | -0.1677 | No |
| 44 | Idh1 | 10956 | -0.069 | -0.1863 | No |
| 45 | Slc25a17 | 11022 | -0.072 | -0.1865 | No |
| 46 | Ech1 | 11360 | -0.087 | -0.2001 | No |
| 47 | Lonp2 | 11706 | -0.103 | -0.2135 | No |
| 48 | Acaa1a | 12466 | -0.135 | -0.2470 | No |
| 49 | Ywhah | 12545 | -0.138 | -0.2449 | No |
| 50 | Abcc5 | 12818 | -0.150 | -0.2524 | No |
| 51 | Ctbp1 | 12834 | -0.151 | -0.2465 | No |
| 52 | Pabpc1 | 12844 | -0.151 | -0.2402 | No |
| 53 | Abcd2 | 12976 | -0.157 | -0.2401 | No |
| 54 | Scp2 | 13165 | -0.165 | -0.2425 | No |
| 55 | Abcd1 | 13507 | -0.179 | -0.2523 | No |
| 56 | Hsd17b4 | 13614 | -0.183 | -0.2497 | No |
| 57 | Eci2 | 13788 | -0.191 | -0.2502 | No |
| 58 | Pex11a | 13893 | -0.196 | -0.2469 | No |
| 59 | Pex14 | 14314 | -0.215 | -0.2592 | No |
| 60 | Ercc1 | 14330 | -0.216 | -0.2504 | No |
| 61 | Ephx2 | 14355 | -0.217 | -0.2420 | No |
| 62 | Atxn1 | 14416 | -0.221 | -0.2353 | No |
| 63 | Pex11b | 15462 | -0.276 | -0.2774 | Yes |
| 64 | Sod2 | 15468 | -0.277 | -0.2654 | Yes |
| 65 | Retsat | 15504 | -0.279 | -0.2548 | Yes |
| 66 | Cdk7 | 15710 | -0.291 | -0.2526 | Yes |
| 67 | Prdx1 | 15714 | -0.291 | -0.2398 | Yes |
| 68 | Pex13 | 15811 | -0.297 | -0.2317 | Yes |
| 69 | Isoc1 | 15933 | -0.306 | -0.2244 | Yes |
| 70 | Mlycd | 16244 | -0.326 | -0.2260 | Yes |
| 71 | Idi1 | 16417 | -0.337 | -0.2200 | Yes |
| 72 | Abcb9 | 16618 | -0.352 | -0.2148 | Yes |
| 73 | Ehhadh | 16710 | -0.359 | -0.2036 | Yes |
| 74 | Cln6 | 16720 | -0.360 | -0.1881 | Yes |
| 75 | Bcl10 | 16795 | -0.365 | -0.1757 | Yes |
| 76 | Gstk1 | 16976 | -0.379 | -0.1682 | Yes |
| 77 | Sult2b1 | 17033 | -0.384 | -0.1541 | Yes |
| 78 | Sod1 | 17704 | -0.442 | -0.1693 | Yes |
| 79 | Dlg4 | 17894 | -0.466 | -0.1584 | Yes |
| 80 | Hsd17b11 | 17992 | -0.483 | -0.1421 | Yes |
| 81 | Prdx5 | 18098 | -0.497 | -0.1255 | Yes |
| 82 | Tspo | 18289 | -0.528 | -0.1119 | Yes |
| 83 | Slc25a4 | 18589 | -0.584 | -0.1015 | Yes |
| 84 | Rdh11 | 18650 | -0.600 | -0.0780 | Yes |
| 85 | Nudt19 | 18723 | -0.618 | -0.0543 | Yes |
| 86 | Cat | 19042 | -0.748 | -0.0376 | Yes |
| 87 | Hras | 19294 | -1.156 | 0.0007 | Yes |

