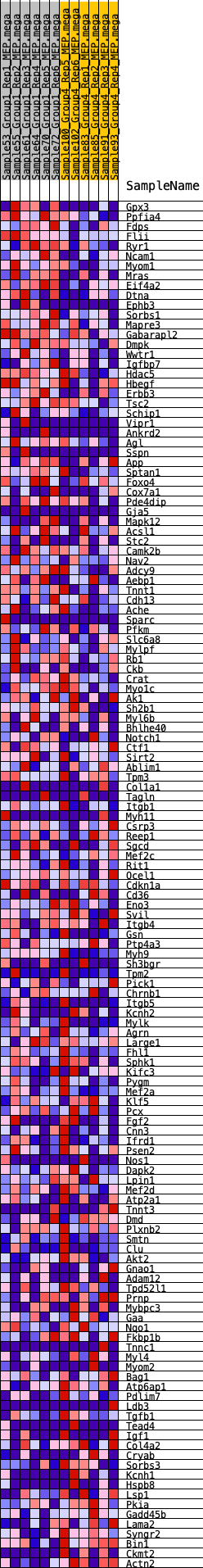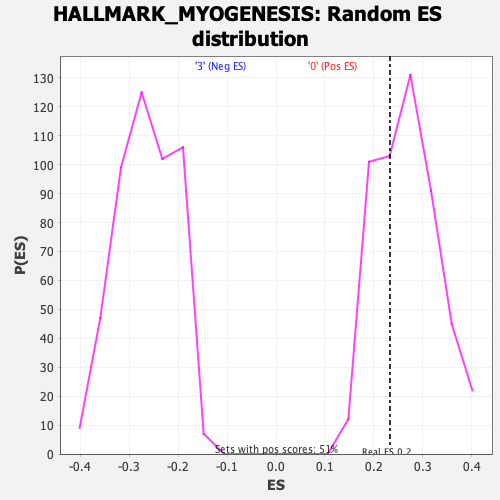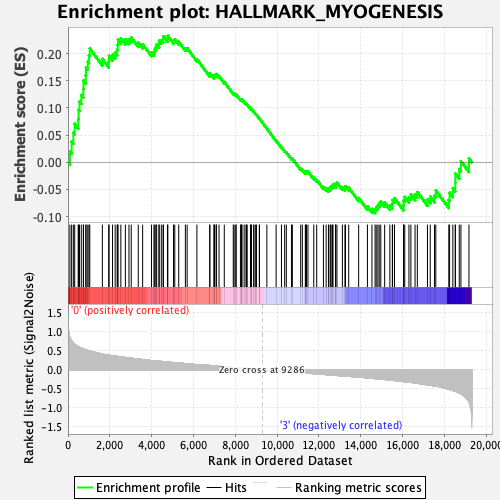
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_MYOGENESIS |
| Enrichment Score (ES) | 0.23279326 |
| Normalized Enrichment Score (NES) | 0.8665819 |
| Nominal p-value | 0.6792079 |
| FDR q-value | 0.8878225 |
| FWER p-Value | 0.998 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Gpx3 | 91 | 0.847 | 0.0200 | Yes |
| 2 | Ppfia4 | 174 | 0.766 | 0.0382 | Yes |
| 3 | Fdps | 255 | 0.705 | 0.0546 | Yes |
| 4 | Flii | 320 | 0.671 | 0.0709 | Yes |
| 5 | Ryr1 | 487 | 0.608 | 0.0800 | Yes |
| 6 | Ncam1 | 507 | 0.603 | 0.0966 | Yes |
| 7 | Myom1 | 549 | 0.591 | 0.1118 | Yes |
| 8 | Mras | 635 | 0.571 | 0.1240 | Yes |
| 9 | Eif4a2 | 733 | 0.546 | 0.1350 | Yes |
| 10 | Dtna | 739 | 0.546 | 0.1507 | Yes |
| 11 | Ephb3 | 850 | 0.524 | 0.1602 | Yes |
| 12 | Sorbs1 | 863 | 0.522 | 0.1749 | Yes |
| 13 | Mapre3 | 945 | 0.508 | 0.1855 | Yes |
| 14 | Gabarapl2 | 1002 | 0.497 | 0.1971 | Yes |
| 15 | Dmpk | 1041 | 0.492 | 0.2095 | Yes |
| 16 | Wwtr1 | 1643 | 0.414 | 0.1903 | Yes |
| 17 | Igfbp7 | 1948 | 0.382 | 0.1856 | Yes |
| 18 | Hdac5 | 1962 | 0.381 | 0.1961 | Yes |
| 19 | Hbegf | 2129 | 0.365 | 0.1981 | Yes |
| 20 | Erbb3 | 2255 | 0.356 | 0.2020 | Yes |
| 21 | Tsc2 | 2349 | 0.350 | 0.2074 | Yes |
| 22 | Schip1 | 2379 | 0.348 | 0.2160 | Yes |
| 23 | Vipr1 | 2389 | 0.347 | 0.2257 | Yes |
| 24 | Ankrd2 | 2527 | 0.335 | 0.2284 | Yes |
| 25 | Agl | 2743 | 0.322 | 0.2266 | Yes |
| 26 | Sspn | 2915 | 0.307 | 0.2266 | Yes |
| 27 | App | 3023 | 0.302 | 0.2299 | Yes |
| 28 | Sptan1 | 3359 | 0.278 | 0.2205 | Yes |
| 29 | Foxo4 | 3569 | 0.266 | 0.2174 | Yes |
| 30 | Cox7a1 | 3991 | 0.240 | 0.2024 | Yes |
| 31 | Pde4dip | 4110 | 0.233 | 0.2031 | Yes |
| 32 | Gja5 | 4149 | 0.232 | 0.2079 | Yes |
| 33 | Mapk12 | 4201 | 0.230 | 0.2120 | Yes |
| 34 | Acsl1 | 4239 | 0.229 | 0.2167 | Yes |
| 35 | Stc2 | 4354 | 0.224 | 0.2173 | Yes |
| 36 | Camk2b | 4355 | 0.224 | 0.2239 | Yes |
| 37 | Nav2 | 4461 | 0.218 | 0.2248 | Yes |
| 38 | Adcy9 | 4546 | 0.212 | 0.2266 | Yes |
| 39 | Aebp1 | 4562 | 0.211 | 0.2320 | Yes |
| 40 | Tnnt1 | 4755 | 0.202 | 0.2279 | Yes |
| 41 | Cdh13 | 4774 | 0.201 | 0.2328 | Yes |
| 42 | Ache | 5045 | 0.188 | 0.2242 | No |
| 43 | Sparc | 5105 | 0.185 | 0.2265 | No |
| 44 | Pfkm | 5294 | 0.177 | 0.2219 | No |
| 45 | Slc6a8 | 5612 | 0.160 | 0.2100 | No |
| 46 | Mylpf | 5696 | 0.155 | 0.2103 | No |
| 47 | Rb1 | 6163 | 0.132 | 0.1898 | No |
| 48 | Ckb | 6778 | 0.111 | 0.1610 | No |
| 49 | Crat | 6781 | 0.111 | 0.1641 | No |
| 50 | Myo1c | 6977 | 0.101 | 0.1569 | No |
| 51 | Ak1 | 6992 | 0.101 | 0.1591 | No |
| 52 | Sh2b1 | 7015 | 0.099 | 0.1609 | No |
| 53 | Myl6b | 7088 | 0.096 | 0.1600 | No |
| 54 | Bhlhe40 | 7094 | 0.096 | 0.1625 | No |
| 55 | Notch1 | 7221 | 0.090 | 0.1586 | No |
| 56 | Ctf1 | 7472 | 0.079 | 0.1478 | No |
| 57 | Sirt2 | 7901 | 0.058 | 0.1272 | No |
| 58 | Ablim1 | 7943 | 0.056 | 0.1267 | No |
| 59 | Tpm3 | 8022 | 0.053 | 0.1242 | No |
| 60 | Col1a1 | 8040 | 0.052 | 0.1248 | No |
| 61 | Tagln | 8245 | 0.042 | 0.1154 | No |
| 62 | Itgb1 | 8292 | 0.041 | 0.1142 | No |
| 63 | Myh11 | 8298 | 0.041 | 0.1151 | No |
| 64 | Csrp3 | 8321 | 0.040 | 0.1151 | No |
| 65 | Reep1 | 8421 | 0.037 | 0.1111 | No |
| 66 | Sgcd | 8510 | 0.033 | 0.1074 | No |
| 67 | Mef2c | 8531 | 0.032 | 0.1073 | No |
| 68 | Rit1 | 8567 | 0.032 | 0.1064 | No |
| 69 | Ocel1 | 8725 | 0.025 | 0.0990 | No |
| 70 | Cdkn1a | 8757 | 0.023 | 0.0980 | No |
| 71 | Cd36 | 8771 | 0.023 | 0.0980 | No |
| 72 | Eno3 | 8874 | 0.018 | 0.0932 | No |
| 73 | Svil | 8940 | 0.015 | 0.0903 | No |
| 74 | Itgb4 | 8981 | 0.013 | 0.0886 | No |
| 75 | Gsn | 9010 | 0.013 | 0.0875 | No |
| 76 | Ptp4a3 | 9147 | 0.006 | 0.0806 | No |
| 77 | Myh9 | 9156 | 0.005 | 0.0803 | No |
| 78 | Sh3bgr | 9508 | -0.004 | 0.0621 | No |
| 79 | Tpm2 | 9951 | -0.024 | 0.0397 | No |
| 80 | Pick1 | 10206 | -0.037 | 0.0276 | No |
| 81 | Chrnb1 | 10360 | -0.042 | 0.0208 | No |
| 82 | Itgb5 | 10438 | -0.046 | 0.0181 | No |
| 83 | Kcnh2 | 10684 | -0.056 | 0.0070 | No |
| 84 | Mylk | 10727 | -0.059 | 0.0065 | No |
| 85 | Agrn | 11126 | -0.076 | -0.0120 | No |
| 86 | Large1 | 11204 | -0.080 | -0.0137 | No |
| 87 | Fhl1 | 11347 | -0.087 | -0.0185 | No |
| 88 | Sphk1 | 11361 | -0.087 | -0.0167 | No |
| 89 | Kifc3 | 11402 | -0.089 | -0.0161 | No |
| 90 | Pygm | 11473 | -0.092 | -0.0171 | No |
| 91 | Mef2a | 11751 | -0.105 | -0.0285 | No |
| 92 | Klf5 | 11891 | -0.112 | -0.0324 | No |
| 93 | Pcx | 12213 | -0.125 | -0.0455 | No |
| 94 | Fgf2 | 12342 | -0.130 | -0.0484 | No |
| 95 | Cnn3 | 12453 | -0.134 | -0.0502 | No |
| 96 | Ifrd1 | 12475 | -0.135 | -0.0474 | No |
| 97 | Psen2 | 12558 | -0.139 | -0.0476 | No |
| 98 | Nos1 | 12579 | -0.140 | -0.0446 | No |
| 99 | Dapk2 | 12648 | -0.143 | -0.0439 | No |
| 100 | Lpin1 | 12670 | -0.144 | -0.0408 | No |
| 101 | Mef2d | 12792 | -0.149 | -0.0428 | No |
| 102 | Atp2a1 | 12798 | -0.149 | -0.0387 | No |
| 103 | Tnnt3 | 12859 | -0.152 | -0.0374 | No |
| 104 | Dmd | 13119 | -0.163 | -0.0461 | No |
| 105 | Plxnb2 | 13241 | -0.169 | -0.0475 | No |
| 106 | Smtn | 13268 | -0.169 | -0.0439 | No |
| 107 | Clu | 13421 | -0.175 | -0.0467 | No |
| 108 | Akt2 | 13897 | -0.197 | -0.0658 | No |
| 109 | Gnao1 | 14316 | -0.215 | -0.0813 | No |
| 110 | Adam12 | 14533 | -0.227 | -0.0859 | No |
| 111 | Tpd52l1 | 14682 | -0.235 | -0.0868 | No |
| 112 | Prnp | 14736 | -0.238 | -0.0826 | No |
| 113 | Mybpc3 | 14816 | -0.243 | -0.0796 | No |
| 114 | Gaa | 14879 | -0.246 | -0.0756 | No |
| 115 | Nqo1 | 14954 | -0.248 | -0.0722 | No |
| 116 | Fkbp1b | 15140 | -0.259 | -0.0743 | No |
| 117 | Tnnc1 | 15389 | -0.274 | -0.0793 | No |
| 118 | Myl4 | 15507 | -0.279 | -0.0772 | No |
| 119 | Myom2 | 15518 | -0.280 | -0.0695 | No |
| 120 | Bag1 | 15611 | -0.286 | -0.0660 | No |
| 121 | Atp6ap1 | 16042 | -0.313 | -0.0793 | No |
| 122 | Pdlim7 | 16045 | -0.313 | -0.0702 | No |
| 123 | Ldb3 | 16100 | -0.317 | -0.0637 | No |
| 124 | Tgfb1 | 16301 | -0.329 | -0.0646 | No |
| 125 | Tead4 | 16394 | -0.336 | -0.0595 | No |
| 126 | Igf1 | 16593 | -0.350 | -0.0596 | No |
| 127 | Col4a2 | 16703 | -0.358 | -0.0548 | No |
| 128 | Cryab | 17187 | -0.398 | -0.0684 | No |
| 129 | Sorbs3 | 17320 | -0.411 | -0.0633 | No |
| 130 | Kcnh1 | 17524 | -0.427 | -0.0614 | No |
| 131 | Hspb8 | 17585 | -0.432 | -0.0518 | No |
| 132 | Lsp1 | 18207 | -0.512 | -0.0693 | No |
| 133 | Pkia | 18245 | -0.521 | -0.0560 | No |
| 134 | Gadd45b | 18400 | -0.548 | -0.0480 | No |
| 135 | Lama2 | 18516 | -0.570 | -0.0373 | No |
| 136 | Syngr2 | 18520 | -0.572 | -0.0208 | No |
| 137 | Bin1 | 18707 | -0.615 | -0.0125 | No |
| 138 | Ckmt2 | 18787 | -0.640 | 0.0021 | No |
| 139 | Actn2 | 19171 | -0.854 | 0.0071 | No |