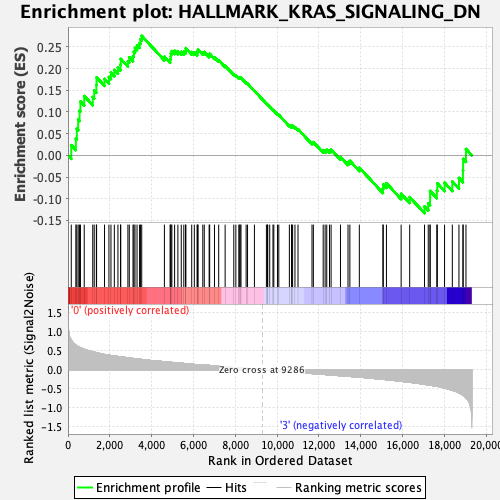
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_KRAS_SIGNALING_DN |
| Enrichment Score (ES) | 0.27521783 |
| Normalized Enrichment Score (NES) | 1.1007975 |
| Nominal p-value | 0.28747433 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.961 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Efhd1 | 156 | 0.783 | 0.0235 | Yes |
| 2 | Atp4a | 375 | 0.644 | 0.0382 | Yes |
| 3 | Clstn3 | 421 | 0.629 | 0.0612 | Yes |
| 4 | Ryr1 | 487 | 0.608 | 0.0824 | Yes |
| 5 | Gpr19 | 552 | 0.591 | 0.1030 | Yes |
| 6 | Prodh | 596 | 0.580 | 0.1242 | Yes |
| 7 | Hsd11b2 | 776 | 0.536 | 0.1365 | Yes |
| 8 | Tfcp2l1 | 1184 | 0.471 | 0.1344 | Yes |
| 9 | Synpo | 1253 | 0.461 | 0.1494 | Yes |
| 10 | Tnni3 | 1356 | 0.446 | 0.1622 | Yes |
| 11 | Skil | 1367 | 0.445 | 0.1796 | Yes |
| 12 | Gtf3c5 | 1746 | 0.400 | 0.1761 | Yes |
| 13 | Coq8a | 1957 | 0.382 | 0.1806 | Yes |
| 14 | Tent5c | 2054 | 0.371 | 0.1906 | Yes |
| 15 | Htr1d | 2215 | 0.360 | 0.1968 | Yes |
| 16 | Slc12a3 | 2388 | 0.347 | 0.2019 | Yes |
| 17 | Epha5 | 2510 | 0.336 | 0.2091 | Yes |
| 18 | Kcnn1 | 2523 | 0.335 | 0.2220 | Yes |
| 19 | Dcc | 2864 | 0.311 | 0.2169 | Yes |
| 20 | Cpa2 | 2930 | 0.307 | 0.2259 | Yes |
| 21 | Mx2 | 3108 | 0.296 | 0.2287 | Yes |
| 22 | Cyp39a1 | 3149 | 0.293 | 0.2384 | Yes |
| 23 | Macroh2a2 | 3202 | 0.290 | 0.2474 | Yes |
| 24 | Tgfb2 | 3301 | 0.281 | 0.2537 | Yes |
| 25 | Pde6b | 3422 | 0.275 | 0.2586 | Yes |
| 26 | Vps50 | 3464 | 0.272 | 0.2675 | Yes |
| 27 | Thrb | 3525 | 0.270 | 0.2752 | Yes |
| 28 | Tgm1 | 4609 | 0.209 | 0.2273 | No |
| 29 | Zc2hc1c | 4881 | 0.196 | 0.2211 | No |
| 30 | Htr1b | 4913 | 0.194 | 0.2273 | No |
| 31 | Tg | 4919 | 0.194 | 0.2348 | No |
| 32 | Kmt2d | 4966 | 0.191 | 0.2401 | No |
| 33 | Zfp112 | 5097 | 0.185 | 0.2409 | No |
| 34 | Abcg4 | 5254 | 0.179 | 0.2400 | No |
| 35 | Fggy | 5415 | 0.171 | 0.2385 | No |
| 36 | Ryr2 | 5537 | 0.164 | 0.2389 | No |
| 37 | Slc16a7 | 5623 | 0.159 | 0.2409 | No |
| 38 | Camk1d | 5633 | 0.159 | 0.2468 | No |
| 39 | Copz2 | 5915 | 0.144 | 0.2380 | No |
| 40 | Tex15 | 6040 | 0.138 | 0.2371 | No |
| 41 | Ngb | 6171 | 0.131 | 0.2356 | No |
| 42 | Arpp21 | 6194 | 0.131 | 0.2398 | No |
| 43 | Cd80 | 6220 | 0.130 | 0.2437 | No |
| 44 | Idua | 6445 | 0.121 | 0.2369 | No |
| 45 | Lgals7 | 6515 | 0.120 | 0.2382 | No |
| 46 | Asb7 | 6748 | 0.113 | 0.2306 | No |
| 47 | Tcf7l1 | 6774 | 0.111 | 0.2338 | No |
| 48 | Msh5 | 7003 | 0.100 | 0.2260 | No |
| 49 | Stag3 | 7205 | 0.090 | 0.2192 | No |
| 50 | Zbtb16 | 7513 | 0.077 | 0.2063 | No |
| 51 | P2rx6 | 7926 | 0.057 | 0.1872 | No |
| 52 | Nr6a1 | 8025 | 0.053 | 0.1842 | No |
| 53 | Plag1 | 8167 | 0.047 | 0.1788 | No |
| 54 | Slc29a3 | 8213 | 0.045 | 0.1782 | No |
| 55 | Sphk2 | 8227 | 0.044 | 0.1793 | No |
| 56 | Nr4a2 | 8271 | 0.042 | 0.1788 | No |
| 57 | Celsr2 | 8517 | 0.033 | 0.1673 | No |
| 58 | Kcnd1 | 8584 | 0.030 | 0.1651 | No |
| 59 | Cpeb3 | 8913 | 0.017 | 0.1487 | No |
| 60 | Ypel1 | 9486 | -0.003 | 0.1191 | No |
| 61 | Prkn | 9539 | -0.005 | 0.1166 | No |
| 62 | Bard1 | 9550 | -0.005 | 0.1163 | No |
| 63 | Ybx2 | 9645 | -0.009 | 0.1118 | No |
| 64 | Cdkal1 | 9800 | -0.016 | 0.1044 | No |
| 65 | Mast3 | 9841 | -0.019 | 0.1031 | No |
| 66 | Btg2 | 10015 | -0.028 | 0.0952 | No |
| 67 | Sgk1 | 10079 | -0.031 | 0.0932 | No |
| 68 | Nrip2 | 10584 | -0.052 | 0.0690 | No |
| 69 | Dtnb | 10690 | -0.057 | 0.0659 | No |
| 70 | Slc6a3 | 10696 | -0.057 | 0.0679 | No |
| 71 | Brdt | 10739 | -0.059 | 0.0681 | No |
| 72 | Ptprj | 10851 | -0.065 | 0.0650 | No |
| 73 | Sptbn2 | 10997 | -0.071 | 0.0603 | No |
| 74 | Gp1ba | 11666 | -0.102 | 0.0296 | No |
| 75 | Itgb1bp2 | 11731 | -0.104 | 0.0305 | No |
| 76 | Bmpr1b | 12201 | -0.124 | 0.0111 | No |
| 77 | Rsad2 | 12293 | -0.128 | 0.0115 | No |
| 78 | Egf | 12371 | -0.131 | 0.0128 | No |
| 79 | Pdk2 | 12503 | -0.137 | 0.0115 | No |
| 80 | Nos1 | 12579 | -0.140 | 0.0132 | No |
| 81 | Serpinb2 | 13028 | -0.159 | -0.0037 | No |
| 82 | Capn9 | 13388 | -0.173 | -0.0154 | No |
| 83 | Entpd7 | 13477 | -0.177 | -0.0128 | No |
| 84 | Grid2 | 13929 | -0.198 | -0.0283 | No |
| 85 | Myo15a | 15056 | -0.255 | -0.0766 | No |
| 86 | Gamt | 15073 | -0.256 | -0.0671 | No |
| 87 | Thnsl2 | 15225 | -0.265 | -0.0642 | No |
| 88 | Selenop | 15928 | -0.305 | -0.0885 | No |
| 89 | Mthfr | 16337 | -0.333 | -0.0963 | No |
| 90 | Col2a1 | 17046 | -0.386 | -0.1175 | No |
| 91 | Lfng | 17224 | -0.401 | -0.1105 | No |
| 92 | Edar | 17309 | -0.409 | -0.0984 | No |
| 93 | Mefv | 17312 | -0.410 | -0.0819 | No |
| 94 | Slc25a23 | 17633 | -0.435 | -0.0810 | No |
| 95 | Tenm2 | 17658 | -0.438 | -0.0646 | No |
| 96 | Mfsd6 | 18004 | -0.484 | -0.0630 | No |
| 97 | Fgfr3 | 18375 | -0.543 | -0.0603 | No |
| 98 | Snn | 18692 | -0.611 | -0.0521 | No |
| 99 | Sidt1 | 18883 | -0.668 | -0.0350 | No |
| 100 | Ccdc106 | 18892 | -0.671 | -0.0083 | No |
| 101 | Klk8 | 19027 | -0.738 | 0.0146 | No |

