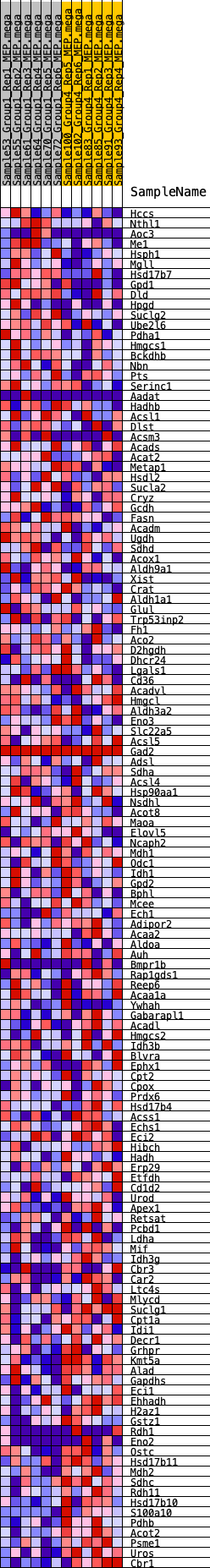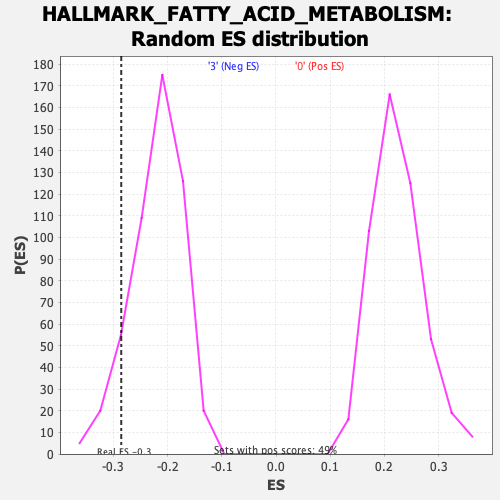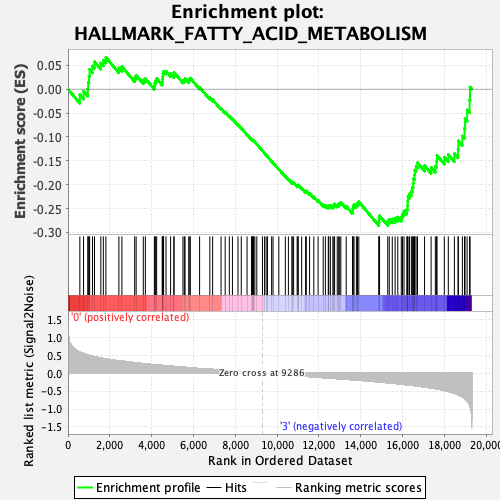
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 3 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.28563264 |
| Normalized Enrichment Score (NES) | -1.3035294 |
| Nominal p-value | 0.1 |
| FDR q-value | 0.5094155 |
| FWER p-Value | 0.763 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Hccs | 567 | 0.586 | -0.0118 | No |
| 2 | Nthl1 | 744 | 0.544 | -0.0045 | No |
| 3 | Aoc3 | 949 | 0.507 | 0.0003 | No |
| 4 | Me1 | 965 | 0.504 | 0.0148 | No |
| 5 | Hsph1 | 1006 | 0.497 | 0.0277 | No |
| 6 | Mgll | 1035 | 0.493 | 0.0412 | No |
| 7 | Hsd17b7 | 1173 | 0.475 | 0.0485 | No |
| 8 | Gpd1 | 1267 | 0.460 | 0.0576 | No |
| 9 | Dld | 1570 | 0.423 | 0.0546 | No |
| 10 | Hpgd | 1689 | 0.405 | 0.0608 | No |
| 11 | Suclg2 | 1811 | 0.394 | 0.0664 | No |
| 12 | Ube2l6 | 2433 | 0.343 | 0.0444 | No |
| 13 | Pdha1 | 2577 | 0.333 | 0.0471 | No |
| 14 | Hmgcs1 | 3188 | 0.291 | 0.0241 | No |
| 15 | Bckdhb | 3258 | 0.286 | 0.0291 | No |
| 16 | Nbn | 3599 | 0.264 | 0.0194 | No |
| 17 | Pts | 3698 | 0.257 | 0.0221 | No |
| 18 | Serinc1 | 4130 | 0.232 | 0.0067 | No |
| 19 | Aadat | 4150 | 0.232 | 0.0127 | No |
| 20 | Hadhb | 4191 | 0.231 | 0.0176 | No |
| 21 | Acsl1 | 4239 | 0.229 | 0.0221 | No |
| 22 | Dlst | 4504 | 0.216 | 0.0149 | No |
| 23 | Acsm3 | 4518 | 0.215 | 0.0207 | No |
| 24 | Acads | 4531 | 0.214 | 0.0266 | No |
| 25 | Acat2 | 4540 | 0.213 | 0.0326 | No |
| 26 | Metap1 | 4570 | 0.211 | 0.0375 | No |
| 27 | Hsdl2 | 4686 | 0.204 | 0.0377 | No |
| 28 | Sucla2 | 4905 | 0.194 | 0.0322 | No |
| 29 | Cryz | 5064 | 0.186 | 0.0296 | No |
| 30 | Gcdh | 5066 | 0.186 | 0.0352 | No |
| 31 | Fasn | 5497 | 0.166 | 0.0178 | No |
| 32 | Acadm | 5562 | 0.163 | 0.0194 | No |
| 33 | Ugdh | 5597 | 0.161 | 0.0225 | No |
| 34 | Sdhd | 5776 | 0.151 | 0.0178 | No |
| 35 | Acox1 | 5790 | 0.150 | 0.0217 | No |
| 36 | Aldh9a1 | 5850 | 0.147 | 0.0231 | No |
| 37 | Xist | 6292 | 0.126 | 0.0039 | No |
| 38 | Crat | 6781 | 0.111 | -0.0182 | No |
| 39 | Aldh1a1 | 6915 | 0.104 | -0.0219 | No |
| 40 | Glul | 7318 | 0.085 | -0.0403 | No |
| 41 | Trp53inp2 | 7515 | 0.077 | -0.0482 | No |
| 42 | Fh1 | 7713 | 0.067 | -0.0564 | No |
| 43 | Aco2 | 7862 | 0.060 | -0.0623 | No |
| 44 | D2hgdh | 8127 | 0.049 | -0.0746 | No |
| 45 | Dhcr24 | 8284 | 0.041 | -0.0815 | No |
| 46 | Lgals1 | 8561 | 0.032 | -0.0949 | No |
| 47 | Cd36 | 8771 | 0.023 | -0.1052 | No |
| 48 | Acadvl | 8814 | 0.021 | -0.1067 | No |
| 49 | Hmgcl | 8822 | 0.020 | -0.1065 | No |
| 50 | Aldh3a2 | 8873 | 0.018 | -0.1085 | No |
| 51 | Eno3 | 8874 | 0.018 | -0.1080 | No |
| 52 | Slc22a5 | 8934 | 0.016 | -0.1106 | No |
| 53 | Acsl5 | 9027 | 0.012 | -0.1150 | No |
| 54 | Gad2 | 9298 | 0.000 | -0.1291 | No |
| 55 | Adsl | 9399 | -0.000 | -0.1343 | No |
| 56 | Sdha | 9473 | -0.003 | -0.1380 | No |
| 57 | Acsl4 | 9536 | -0.005 | -0.1411 | No |
| 58 | Hsp90aa1 | 9736 | -0.013 | -0.1511 | No |
| 59 | Nsdhl | 9803 | -0.017 | -0.1540 | No |
| 60 | Acot8 | 10080 | -0.031 | -0.1675 | No |
| 61 | Maoa | 10391 | -0.044 | -0.1823 | No |
| 62 | Elovl5 | 10538 | -0.050 | -0.1884 | No |
| 63 | Ncaph2 | 10708 | -0.058 | -0.1955 | No |
| 64 | Mdh1 | 10721 | -0.058 | -0.1943 | No |
| 65 | Odc1 | 10787 | -0.062 | -0.1958 | No |
| 66 | Idh1 | 10956 | -0.069 | -0.2025 | No |
| 67 | Gpd2 | 10999 | -0.071 | -0.2025 | No |
| 68 | Bphl | 11002 | -0.071 | -0.2005 | No |
| 69 | Mcee | 11173 | -0.078 | -0.2070 | No |
| 70 | Ech1 | 11360 | -0.087 | -0.2140 | No |
| 71 | Adipor2 | 11393 | -0.089 | -0.2130 | No |
| 72 | Acaa2 | 11551 | -0.096 | -0.2183 | No |
| 73 | Aldoa | 11753 | -0.105 | -0.2255 | No |
| 74 | Auh | 11958 | -0.114 | -0.2327 | No |
| 75 | Bmpr1b | 12201 | -0.124 | -0.2416 | No |
| 76 | Rap1gds1 | 12306 | -0.128 | -0.2431 | No |
| 77 | Reep6 | 12444 | -0.134 | -0.2462 | No |
| 78 | Acaa1a | 12466 | -0.135 | -0.2432 | No |
| 79 | Ywhah | 12545 | -0.138 | -0.2431 | No |
| 80 | Gabarapl1 | 12664 | -0.143 | -0.2449 | No |
| 81 | Acadl | 12712 | -0.145 | -0.2429 | No |
| 82 | Hmgcs2 | 12746 | -0.147 | -0.2402 | No |
| 83 | Idh3b | 12873 | -0.153 | -0.2422 | No |
| 84 | Blvra | 12941 | -0.156 | -0.2409 | No |
| 85 | Ephx1 | 12990 | -0.157 | -0.2387 | No |
| 86 | Cpt2 | 13045 | -0.160 | -0.2366 | No |
| 87 | Cpox | 13302 | -0.171 | -0.2448 | No |
| 88 | Prdx6 | 13601 | -0.183 | -0.2548 | No |
| 89 | Hsd17b4 | 13614 | -0.183 | -0.2498 | No |
| 90 | Acss1 | 13633 | -0.184 | -0.2452 | No |
| 91 | Echs1 | 13675 | -0.186 | -0.2417 | No |
| 92 | Eci2 | 13788 | -0.191 | -0.2417 | No |
| 93 | Hibch | 13842 | -0.194 | -0.2386 | No |
| 94 | Hadh | 13905 | -0.197 | -0.2359 | No |
| 95 | Erp29 | 14860 | -0.245 | -0.2782 | Yes |
| 96 | Etfdh | 14877 | -0.246 | -0.2716 | Yes |
| 97 | Cd1d2 | 14895 | -0.247 | -0.2650 | Yes |
| 98 | Urod | 15289 | -0.269 | -0.2773 | Yes |
| 99 | Apex1 | 15360 | -0.272 | -0.2727 | Yes |
| 100 | Retsat | 15504 | -0.279 | -0.2717 | Yes |
| 101 | Pcbd1 | 15640 | -0.287 | -0.2700 | Yes |
| 102 | Ldha | 15768 | -0.294 | -0.2678 | Yes |
| 103 | Mif | 15934 | -0.306 | -0.2671 | Yes |
| 104 | Idh3g | 15993 | -0.310 | -0.2607 | Yes |
| 105 | Cbr3 | 16066 | -0.315 | -0.2549 | Yes |
| 106 | Car2 | 16199 | -0.323 | -0.2520 | Yes |
| 107 | Ltc4s | 16232 | -0.325 | -0.2438 | Yes |
| 108 | Mlycd | 16244 | -0.326 | -0.2345 | Yes |
| 109 | Suclg1 | 16253 | -0.326 | -0.2250 | Yes |
| 110 | Cpt1a | 16329 | -0.332 | -0.2189 | Yes |
| 111 | Idi1 | 16417 | -0.337 | -0.2132 | Yes |
| 112 | Decr1 | 16464 | -0.341 | -0.2053 | Yes |
| 113 | Grhpr | 16501 | -0.343 | -0.1967 | Yes |
| 114 | Kmt5a | 16520 | -0.345 | -0.1872 | Yes |
| 115 | Alad | 16568 | -0.348 | -0.1791 | Yes |
| 116 | Gapdhs | 16585 | -0.349 | -0.1693 | Yes |
| 117 | Eci1 | 16643 | -0.354 | -0.1616 | Yes |
| 118 | Ehhadh | 16710 | -0.359 | -0.1541 | Yes |
| 119 | H2az1 | 17047 | -0.386 | -0.1600 | Yes |
| 120 | Gstz1 | 17361 | -0.415 | -0.1637 | Yes |
| 121 | Rdh1 | 17563 | -0.430 | -0.1611 | Yes |
| 122 | Eno2 | 17623 | -0.434 | -0.1510 | Yes |
| 123 | Ostc | 17642 | -0.436 | -0.1388 | Yes |
| 124 | Hsd17b11 | 17992 | -0.483 | -0.1423 | Yes |
| 125 | Mdh2 | 18183 | -0.509 | -0.1368 | Yes |
| 126 | Sdhc | 18473 | -0.562 | -0.1348 | Yes |
| 127 | Rdh11 | 18650 | -0.600 | -0.1258 | Yes |
| 128 | Hsd17b10 | 18663 | -0.604 | -0.1081 | Yes |
| 129 | S100a10 | 18855 | -0.658 | -0.0981 | Yes |
| 130 | Pdhb | 18957 | -0.700 | -0.0822 | Yes |
| 131 | Acot2 | 18983 | -0.713 | -0.0618 | Yes |
| 132 | Psme1 | 19082 | -0.775 | -0.0434 | Yes |
| 133 | Uros | 19202 | -0.891 | -0.0226 | Yes |
| 134 | Cbr1 | 19222 | -0.925 | 0.0044 | Yes |