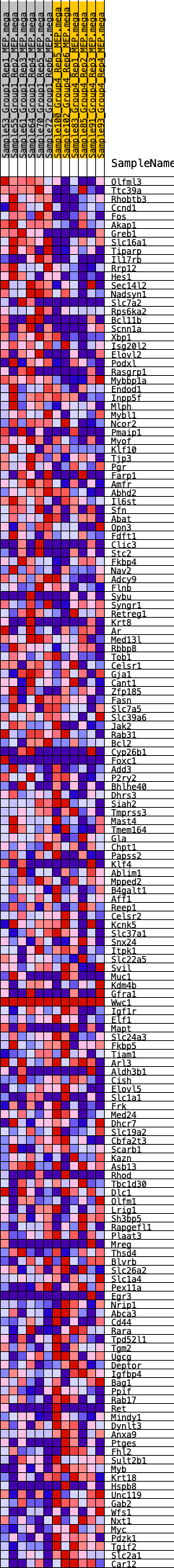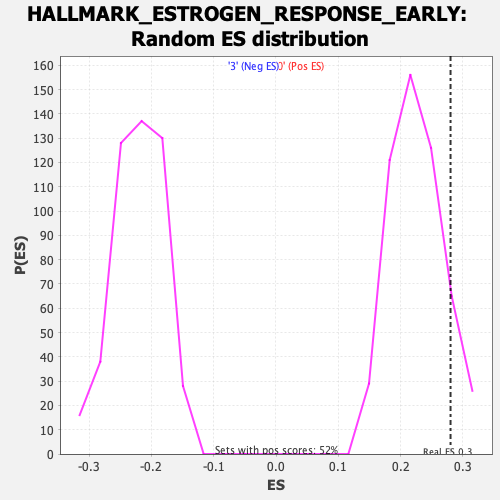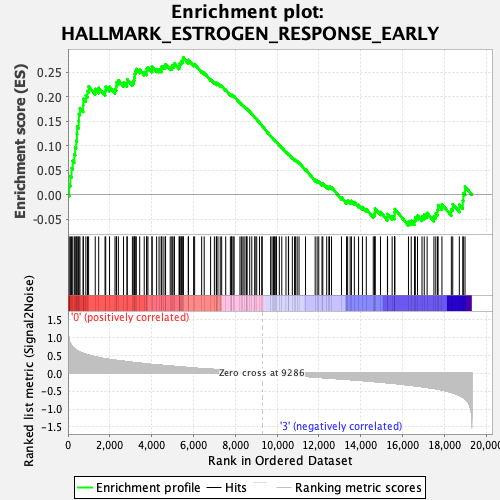
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_ESTROGEN_RESPONSE_EARLY |
| Enrichment Score (ES) | 0.28032 |
| Normalized Enrichment Score (NES) | 1.2476053 |
| Nominal p-value | 0.09751434 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.857 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Olfml3 | 57 | 0.935 | 0.0203 | Yes |
| 2 | Ttc39a | 114 | 0.823 | 0.0379 | Yes |
| 3 | Rhobtb3 | 170 | 0.768 | 0.0542 | Yes |
| 4 | Ccnd1 | 226 | 0.722 | 0.0694 | Yes |
| 5 | Fos | 302 | 0.683 | 0.0825 | Yes |
| 6 | Akap1 | 343 | 0.656 | 0.0968 | Yes |
| 7 | Greb1 | 392 | 0.639 | 0.1102 | Yes |
| 8 | Slc16a1 | 426 | 0.628 | 0.1241 | Yes |
| 9 | Tiparp | 435 | 0.625 | 0.1393 | Yes |
| 10 | Il17rb | 512 | 0.601 | 0.1503 | Yes |
| 11 | Rrp12 | 521 | 0.598 | 0.1648 | Yes |
| 12 | Hes1 | 571 | 0.585 | 0.1768 | Yes |
| 13 | Sec14l2 | 724 | 0.549 | 0.1826 | Yes |
| 14 | Nadsyn1 | 731 | 0.547 | 0.1959 | Yes |
| 15 | Slc7a2 | 854 | 0.524 | 0.2026 | Yes |
| 16 | Rps6ka2 | 932 | 0.509 | 0.2113 | Yes |
| 17 | Bcl11b | 985 | 0.501 | 0.2211 | Yes |
| 18 | Scnn1a | 1302 | 0.455 | 0.2159 | Yes |
| 19 | Xbp1 | 1467 | 0.433 | 0.2181 | Yes |
| 20 | Isg20l2 | 1771 | 0.397 | 0.2122 | Yes |
| 21 | Elovl2 | 1803 | 0.394 | 0.2204 | Yes |
| 22 | Podxl | 1986 | 0.379 | 0.2204 | Yes |
| 23 | Rasgrp1 | 2247 | 0.357 | 0.2157 | Yes |
| 24 | Mybbp1a | 2311 | 0.353 | 0.2212 | Yes |
| 25 | Endod1 | 2320 | 0.352 | 0.2296 | Yes |
| 26 | Inpp5f | 2408 | 0.346 | 0.2337 | Yes |
| 27 | Mlph | 2655 | 0.329 | 0.2290 | Yes |
| 28 | Mybl1 | 2820 | 0.314 | 0.2283 | Yes |
| 29 | Ncor2 | 2832 | 0.313 | 0.2355 | Yes |
| 30 | Pmaip1 | 3079 | 0.297 | 0.2301 | Yes |
| 31 | Myof | 3148 | 0.293 | 0.2339 | Yes |
| 32 | Klf10 | 3170 | 0.292 | 0.2401 | Yes |
| 33 | Tjp3 | 3190 | 0.290 | 0.2463 | Yes |
| 34 | Pgr | 3214 | 0.289 | 0.2523 | Yes |
| 35 | Farp1 | 3266 | 0.285 | 0.2567 | Yes |
| 36 | Amfr | 3428 | 0.275 | 0.2552 | Yes |
| 37 | Abhd2 | 3644 | 0.261 | 0.2505 | Yes |
| 38 | Il6st | 3761 | 0.254 | 0.2507 | Yes |
| 39 | Sfn | 3762 | 0.254 | 0.2571 | Yes |
| 40 | Abat | 3822 | 0.250 | 0.2602 | Yes |
| 41 | Opn3 | 4016 | 0.238 | 0.2561 | Yes |
| 42 | Fdft1 | 4018 | 0.238 | 0.2620 | Yes |
| 43 | Clic3 | 4237 | 0.229 | 0.2563 | Yes |
| 44 | Stc2 | 4354 | 0.224 | 0.2558 | Yes |
| 45 | Fkbp4 | 4460 | 0.218 | 0.2558 | Yes |
| 46 | Nav2 | 4461 | 0.218 | 0.2612 | Yes |
| 47 | Adcy9 | 4546 | 0.212 | 0.2621 | Yes |
| 48 | Flnb | 4642 | 0.207 | 0.2623 | Yes |
| 49 | Sybu | 4665 | 0.205 | 0.2663 | Yes |
| 50 | Syngr1 | 4887 | 0.195 | 0.2596 | Yes |
| 51 | Retreg1 | 4969 | 0.191 | 0.2601 | Yes |
| 52 | Krt8 | 4981 | 0.190 | 0.2643 | Yes |
| 53 | Ar | 5067 | 0.186 | 0.2645 | Yes |
| 54 | Med13l | 5088 | 0.185 | 0.2681 | Yes |
| 55 | Rbbp8 | 5311 | 0.176 | 0.2609 | Yes |
| 56 | Tob1 | 5332 | 0.175 | 0.2642 | Yes |
| 57 | Celsr1 | 5339 | 0.174 | 0.2682 | Yes |
| 58 | Gja1 | 5394 | 0.172 | 0.2697 | Yes |
| 59 | Cant1 | 5422 | 0.170 | 0.2725 | Yes |
| 60 | Zfp185 | 5495 | 0.166 | 0.2729 | Yes |
| 61 | Fasn | 5497 | 0.166 | 0.2770 | Yes |
| 62 | Slc7a5 | 5513 | 0.165 | 0.2803 | Yes |
| 63 | Slc39a6 | 5754 | 0.152 | 0.2716 | No |
| 64 | Jak2 | 5755 | 0.152 | 0.2754 | No |
| 65 | Rab31 | 6006 | 0.139 | 0.2658 | No |
| 66 | Bcl2 | 6055 | 0.137 | 0.2667 | No |
| 67 | Cyp26b1 | 6391 | 0.123 | 0.2523 | No |
| 68 | Foxc1 | 6512 | 0.120 | 0.2490 | No |
| 69 | Add3 | 6818 | 0.109 | 0.2358 | No |
| 70 | P2ry2 | 6998 | 0.100 | 0.2289 | No |
| 71 | Bhlhe40 | 7094 | 0.096 | 0.2264 | No |
| 72 | Dhrs3 | 7095 | 0.096 | 0.2288 | No |
| 73 | Siah2 | 7168 | 0.092 | 0.2273 | No |
| 74 | Tmprss3 | 7278 | 0.087 | 0.2238 | No |
| 75 | Mast4 | 7347 | 0.084 | 0.2223 | No |
| 76 | Tmem164 | 7540 | 0.076 | 0.2142 | No |
| 77 | Gla | 7763 | 0.065 | 0.2042 | No |
| 78 | Chpt1 | 7821 | 0.062 | 0.2028 | No |
| 79 | Papss2 | 7834 | 0.062 | 0.2037 | No |
| 80 | Klf4 | 7896 | 0.058 | 0.2020 | No |
| 81 | Ablim1 | 7943 | 0.056 | 0.2010 | No |
| 82 | Mpped2 | 8222 | 0.044 | 0.1875 | No |
| 83 | B4galt1 | 8289 | 0.041 | 0.1851 | No |
| 84 | Aff1 | 8362 | 0.039 | 0.1823 | No |
| 85 | Reep1 | 8421 | 0.037 | 0.1802 | No |
| 86 | Celsr2 | 8517 | 0.033 | 0.1761 | No |
| 87 | Kcnk5 | 8529 | 0.033 | 0.1763 | No |
| 88 | Slc37a1 | 8564 | 0.032 | 0.1753 | No |
| 89 | Snx24 | 8689 | 0.026 | 0.1695 | No |
| 90 | Itpk1 | 8788 | 0.022 | 0.1649 | No |
| 91 | Slc22a5 | 8934 | 0.016 | 0.1577 | No |
| 92 | Svil | 8940 | 0.015 | 0.1579 | No |
| 93 | Muc1 | 9023 | 0.012 | 0.1539 | No |
| 94 | Kdm4b | 9157 | 0.005 | 0.1471 | No |
| 95 | Gfra1 | 9264 | 0.001 | 0.1416 | No |
| 96 | Wwc1 | 9286 | 0.000 | 0.1405 | No |
| 97 | Igf1r | 9688 | -0.012 | 0.1198 | No |
| 98 | Elf1 | 9756 | -0.014 | 0.1167 | No |
| 99 | Mapt | 9827 | -0.018 | 0.1135 | No |
| 100 | Slc24a3 | 9869 | -0.020 | 0.1118 | No |
| 101 | Fkbp5 | 9932 | -0.023 | 0.1091 | No |
| 102 | Tiam1 | 9970 | -0.025 | 0.1078 | No |
| 103 | Arl3 | 10104 | -0.032 | 0.1017 | No |
| 104 | Aldh3b1 | 10221 | -0.038 | 0.0966 | No |
| 105 | Cish | 10422 | -0.045 | 0.0872 | No |
| 106 | Elovl5 | 10538 | -0.050 | 0.0825 | No |
| 107 | Slc1a1 | 10546 | -0.051 | 0.0834 | No |
| 108 | Frk | 10726 | -0.059 | 0.0755 | No |
| 109 | Med24 | 10832 | -0.064 | 0.0716 | No |
| 110 | Dhcr7 | 10883 | -0.066 | 0.0706 | No |
| 111 | Slc19a2 | 10957 | -0.069 | 0.0686 | No |
| 112 | Cbfa2t3 | 11046 | -0.073 | 0.0658 | No |
| 113 | Scarb1 | 11357 | -0.087 | 0.0518 | No |
| 114 | Kazn | 11817 | -0.108 | 0.0305 | No |
| 115 | Asb13 | 11911 | -0.112 | 0.0284 | No |
| 116 | Rhod | 11989 | -0.115 | 0.0273 | No |
| 117 | Tbc1d30 | 12145 | -0.122 | 0.0222 | No |
| 118 | Dlc1 | 12183 | -0.124 | 0.0234 | No |
| 119 | Olfm1 | 12369 | -0.131 | 0.0170 | No |
| 120 | Lrig1 | 12472 | -0.135 | 0.0150 | No |
| 121 | Sh3bp5 | 12504 | -0.137 | 0.0168 | No |
| 122 | Rapgefl1 | 12600 | -0.140 | 0.0153 | No |
| 123 | Plaat3 | 13075 | -0.161 | -0.0054 | No |
| 124 | Mreg | 13319 | -0.172 | -0.0138 | No |
| 125 | Thsd4 | 13370 | -0.172 | -0.0121 | No |
| 126 | Blvrb | 13491 | -0.178 | -0.0139 | No |
| 127 | Slc26a2 | 13551 | -0.181 | -0.0125 | No |
| 128 | Slc1a4 | 13691 | -0.187 | -0.0151 | No |
| 129 | Pex11a | 13893 | -0.196 | -0.0207 | No |
| 130 | Egr3 | 14078 | -0.206 | -0.0252 | No |
| 131 | Nrip1 | 14262 | -0.212 | -0.0294 | No |
| 132 | Abca3 | 14595 | -0.230 | -0.0410 | No |
| 133 | Cd44 | 14655 | -0.234 | -0.0383 | No |
| 134 | Rara | 14676 | -0.234 | -0.0335 | No |
| 135 | Tpd52l1 | 14682 | -0.235 | -0.0279 | No |
| 136 | Tgm2 | 14940 | -0.247 | -0.0351 | No |
| 137 | Ugcg | 15272 | -0.267 | -0.0458 | No |
| 138 | Deptor | 15279 | -0.268 | -0.0394 | No |
| 139 | Igfbp4 | 15498 | -0.278 | -0.0438 | No |
| 140 | Bag1 | 15611 | -0.286 | -0.0426 | No |
| 141 | Ppif | 15614 | -0.286 | -0.0355 | No |
| 142 | Rab17 | 15629 | -0.287 | -0.0291 | No |
| 143 | Ret | 16278 | -0.329 | -0.0548 | No |
| 144 | Mindy1 | 16410 | -0.337 | -0.0532 | No |
| 145 | Dynlt3 | 16565 | -0.348 | -0.0526 | No |
| 146 | Anxa9 | 16606 | -0.351 | -0.0459 | No |
| 147 | Ptges | 16711 | -0.359 | -0.0424 | No |
| 148 | Fhl2 | 16926 | -0.375 | -0.0442 | No |
| 149 | Sult2b1 | 17033 | -0.384 | -0.0402 | No |
| 150 | Myb | 17170 | -0.397 | -0.0374 | No |
| 151 | Krt18 | 17496 | -0.425 | -0.0437 | No |
| 152 | Hspb8 | 17585 | -0.432 | -0.0375 | No |
| 153 | Unc119 | 17669 | -0.439 | -0.0309 | No |
| 154 | Gab2 | 17694 | -0.441 | -0.0212 | No |
| 155 | Wfs1 | 17879 | -0.464 | -0.0192 | No |
| 156 | Nxt1 | 18328 | -0.535 | -0.0293 | No |
| 157 | Myc | 18395 | -0.548 | -0.0191 | No |
| 158 | Pdzk1 | 18710 | -0.615 | -0.0201 | No |
| 159 | Tgif2 | 18877 | -0.664 | -0.0122 | No |
| 160 | Slc2a1 | 18900 | -0.673 | 0.0034 | No |
| 161 | Car12 | 18982 | -0.713 | 0.0170 | No |