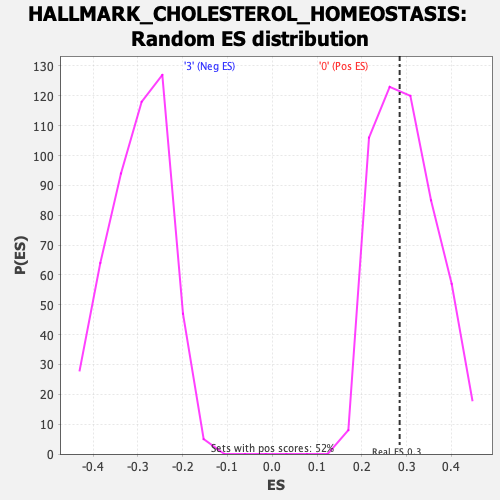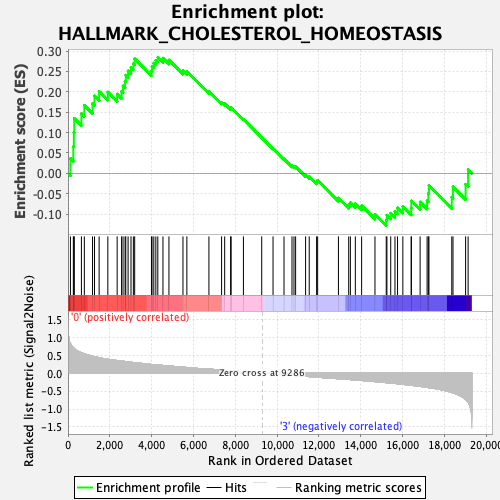
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group4.MEP.mega_Pheno.cls #Group1_versus_Group4_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group4_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_CHOLESTEROL_HOMEOSTASIS |
| Enrichment Score (ES) | 0.28447685 |
| Normalized Enrichment Score (NES) | 0.9565715 |
| Nominal p-value | 0.549323 |
| FDR q-value | 0.8030557 |
| FWER p-Value | 0.99 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pmvk | 116 | 0.821 | 0.0365 | Yes |
| 2 | Fdps | 255 | 0.705 | 0.0658 | Yes |
| 3 | Trp53inp1 | 280 | 0.693 | 0.1004 | Yes |
| 4 | Trib3 | 299 | 0.684 | 0.1349 | Yes |
| 5 | Mvk | 641 | 0.569 | 0.1467 | Yes |
| 6 | Cpeb2 | 781 | 0.535 | 0.1671 | Yes |
| 7 | Hsd17b7 | 1173 | 0.475 | 0.1714 | Yes |
| 8 | Sqle | 1268 | 0.460 | 0.1903 | Yes |
| 9 | Hmgcr | 1488 | 0.431 | 0.2012 | Yes |
| 10 | Lpl | 1905 | 0.385 | 0.1995 | Yes |
| 11 | Atxn2 | 2350 | 0.350 | 0.1946 | Yes |
| 12 | Atf3 | 2561 | 0.334 | 0.2010 | Yes |
| 13 | Gnai1 | 2631 | 0.331 | 0.2145 | Yes |
| 14 | Ldlr | 2728 | 0.324 | 0.2263 | Yes |
| 15 | Cyp51 | 2763 | 0.319 | 0.2410 | Yes |
| 16 | Srebf2 | 2873 | 0.310 | 0.2514 | Yes |
| 17 | Ctnnb1 | 3012 | 0.303 | 0.2600 | Yes |
| 18 | Tm7sf2 | 3125 | 0.295 | 0.2694 | Yes |
| 19 | Hmgcs1 | 3188 | 0.291 | 0.2812 | Yes |
| 20 | Gpx8 | 3990 | 0.240 | 0.2520 | Yes |
| 21 | Fdft1 | 4018 | 0.238 | 0.2629 | Yes |
| 22 | Jag1 | 4103 | 0.234 | 0.2707 | Yes |
| 23 | Pparg | 4200 | 0.230 | 0.2776 | Yes |
| 24 | Gldc | 4294 | 0.226 | 0.2845 | Yes |
| 25 | Acat2 | 4540 | 0.213 | 0.2828 | No |
| 26 | Alcam | 4826 | 0.198 | 0.2782 | No |
| 27 | Fasn | 5497 | 0.166 | 0.2520 | No |
| 28 | Fabp5 | 5683 | 0.156 | 0.2505 | No |
| 29 | Tmem97 | 6738 | 0.113 | 0.2015 | No |
| 30 | Stard4 | 7343 | 0.084 | 0.1745 | No |
| 31 | Chka | 7491 | 0.078 | 0.1709 | No |
| 32 | Fads2 | 7774 | 0.065 | 0.1596 | No |
| 33 | Abca2 | 7799 | 0.063 | 0.1616 | No |
| 34 | Plscr1 | 8387 | 0.038 | 0.1331 | No |
| 35 | Pdk3 | 9262 | 0.001 | 0.0877 | No |
| 36 | Nsdhl | 9803 | -0.017 | 0.0605 | No |
| 37 | Lgals3 | 10328 | -0.042 | 0.0355 | No |
| 38 | Antxr2 | 10709 | -0.058 | 0.0187 | No |
| 39 | Nfil3 | 10805 | -0.063 | 0.0170 | No |
| 40 | Dhcr7 | 10883 | -0.066 | 0.0164 | No |
| 41 | Ech1 | 11360 | -0.087 | -0.0038 | No |
| 42 | Cd9 | 11533 | -0.095 | -0.0078 | No |
| 43 | Anxa5 | 11886 | -0.112 | -0.0203 | No |
| 44 | Acss2 | 11936 | -0.114 | -0.0170 | No |
| 45 | Stx5a | 12930 | -0.155 | -0.0606 | No |
| 46 | Clu | 13421 | -0.175 | -0.0770 | No |
| 47 | Niban1 | 13501 | -0.179 | -0.0718 | No |
| 48 | Pcyt2 | 13738 | -0.189 | -0.0743 | No |
| 49 | Errfi1 | 14039 | -0.204 | -0.0793 | No |
| 50 | Ebp | 14677 | -0.234 | -0.1003 | No |
| 51 | Aldoc | 15215 | -0.264 | -0.1146 | No |
| 52 | Atf5 | 15248 | -0.266 | -0.1025 | No |
| 53 | Ethe1 | 15430 | -0.275 | -0.0976 | No |
| 54 | Sc5d | 15633 | -0.287 | -0.0933 | No |
| 55 | Actg1 | 15756 | -0.293 | -0.0845 | No |
| 56 | S100a11 | 16010 | -0.311 | -0.0815 | No |
| 57 | Lgmn | 16409 | -0.337 | -0.0847 | No |
| 58 | Idi1 | 16417 | -0.337 | -0.0677 | No |
| 59 | Tnfrsf12a | 16835 | -0.369 | -0.0703 | No |
| 60 | Gusb | 17169 | -0.397 | -0.0670 | No |
| 61 | Mvd | 17228 | -0.402 | -0.0492 | No |
| 62 | Pnrc1 | 17258 | -0.405 | -0.0297 | No |
| 63 | Lss | 18349 | -0.539 | -0.0585 | No |
| 64 | Gstm7 | 18407 | -0.549 | -0.0330 | No |
| 65 | Plaur | 19010 | -0.726 | -0.0267 | No |
| 66 | Fbxo6 | 19130 | -0.813 | 0.0092 | No |