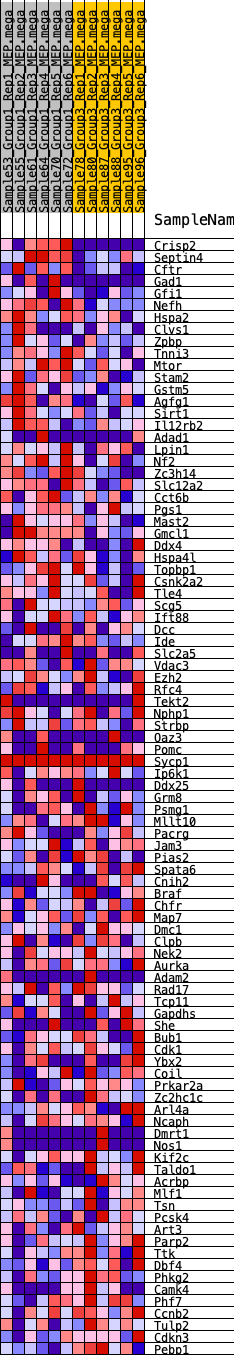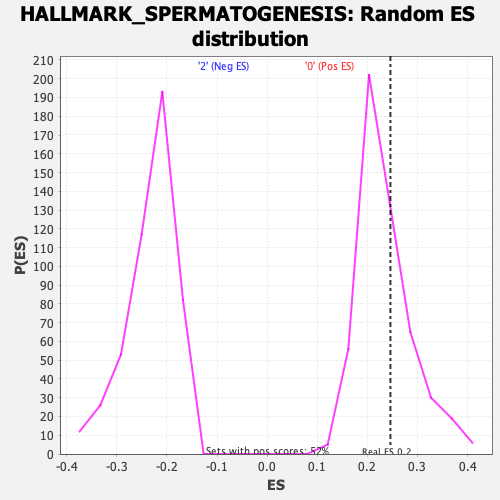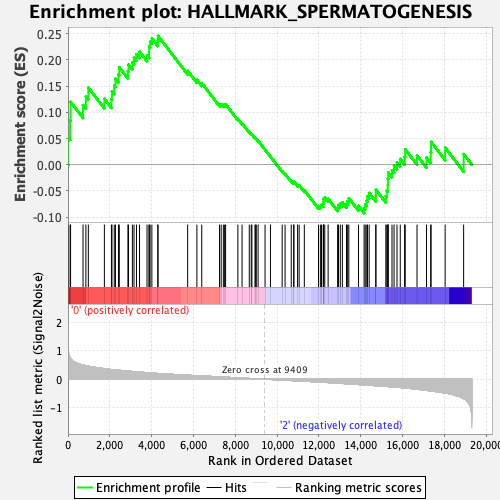
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
| Enrichment Score (ES) | 0.24599615 |
| Normalized Enrichment Score (NES) | 1.0528013 |
| Nominal p-value | 0.34816247 |
| FDR q-value | 0.7695776 |
| FWER p-Value | 0.988 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Crisp2 | 12 | 1.047 | 0.0516 | Yes |
| 2 | Septin4 | 101 | 0.754 | 0.0847 | Yes |
| 3 | Cftr | 123 | 0.729 | 0.1200 | Yes |
| 4 | Gad1 | 718 | 0.495 | 0.1138 | Yes |
| 5 | Gfi1 | 855 | 0.476 | 0.1305 | Yes |
| 6 | Nefh | 972 | 0.456 | 0.1472 | Yes |
| 7 | Hspa2 | 1739 | 0.368 | 0.1257 | Yes |
| 8 | Clvs1 | 2076 | 0.335 | 0.1250 | Yes |
| 9 | Zpbp | 2102 | 0.334 | 0.1404 | Yes |
| 10 | Tnni3 | 2211 | 0.328 | 0.1511 | Yes |
| 11 | Mtor | 2268 | 0.323 | 0.1643 | Yes |
| 12 | Stam2 | 2414 | 0.313 | 0.1724 | Yes |
| 13 | Gstm5 | 2452 | 0.310 | 0.1859 | Yes |
| 14 | Agfg1 | 2865 | 0.283 | 0.1786 | Yes |
| 15 | Sirt1 | 2894 | 0.281 | 0.1912 | Yes |
| 16 | Il12rb2 | 3078 | 0.271 | 0.1952 | Yes |
| 17 | Adad1 | 3156 | 0.265 | 0.2044 | Yes |
| 18 | Lpin1 | 3274 | 0.257 | 0.2111 | Yes |
| 19 | Nf2 | 3420 | 0.248 | 0.2159 | Yes |
| 20 | Zc3h14 | 3778 | 0.230 | 0.2089 | Yes |
| 21 | Slc12a2 | 3878 | 0.224 | 0.2149 | Yes |
| 22 | Cct6b | 3880 | 0.224 | 0.2260 | Yes |
| 23 | Pgs1 | 3926 | 0.221 | 0.2347 | Yes |
| 24 | Mast2 | 4010 | 0.216 | 0.2412 | Yes |
| 25 | Gmcl1 | 4296 | 0.200 | 0.2363 | Yes |
| 26 | Ddx4 | 4303 | 0.200 | 0.2460 | Yes |
| 27 | Hspa4l | 5721 | 0.138 | 0.1792 | No |
| 28 | Topbp1 | 6163 | 0.121 | 0.1622 | No |
| 29 | Csnk2a2 | 6395 | 0.114 | 0.1559 | No |
| 30 | Tle4 | 7245 | 0.081 | 0.1157 | No |
| 31 | Scg5 | 7323 | 0.078 | 0.1156 | No |
| 32 | Ift88 | 7427 | 0.074 | 0.1140 | No |
| 33 | Dcc | 7489 | 0.071 | 0.1143 | No |
| 34 | Ide | 7535 | 0.070 | 0.1155 | No |
| 35 | Slc2a5 | 8122 | 0.048 | 0.0874 | No |
| 36 | Vdac3 | 8322 | 0.040 | 0.0790 | No |
| 37 | Ezh2 | 8667 | 0.026 | 0.0624 | No |
| 38 | Rfc4 | 8760 | 0.022 | 0.0587 | No |
| 39 | Tekt2 | 8792 | 0.021 | 0.0581 | No |
| 40 | Nphp1 | 8941 | 0.016 | 0.0512 | No |
| 41 | Strbp | 8974 | 0.015 | 0.0503 | No |
| 42 | Oaz3 | 9000 | 0.014 | 0.0497 | No |
| 43 | Pomc | 9089 | 0.011 | 0.0457 | No |
| 44 | Sycp1 | 9421 | 0.000 | 0.0285 | No |
| 45 | Ip6k1 | 9680 | -0.008 | 0.0154 | No |
| 46 | Ddx25 | 10240 | -0.029 | -0.0122 | No |
| 47 | Grm8 | 10379 | -0.034 | -0.0177 | No |
| 48 | Psmg1 | 10674 | -0.046 | -0.0307 | No |
| 49 | Mllt10 | 10790 | -0.050 | -0.0342 | No |
| 50 | Pacrg | 10802 | -0.050 | -0.0323 | No |
| 51 | Jam3 | 10978 | -0.057 | -0.0385 | No |
| 52 | Pias2 | 11052 | -0.060 | -0.0393 | No |
| 53 | Spata6 | 11299 | -0.069 | -0.0487 | No |
| 54 | Cnih2 | 11977 | -0.098 | -0.0790 | No |
| 55 | Braf | 12066 | -0.101 | -0.0786 | No |
| 56 | Chfr | 12121 | -0.103 | -0.0762 | No |
| 57 | Map7 | 12204 | -0.107 | -0.0752 | No |
| 58 | Dmc1 | 12216 | -0.107 | -0.0704 | No |
| 59 | Clpb | 12218 | -0.107 | -0.0651 | No |
| 60 | Nek2 | 12276 | -0.110 | -0.0626 | No |
| 61 | Aurka | 12437 | -0.117 | -0.0651 | No |
| 62 | Adam2 | 12891 | -0.137 | -0.0818 | No |
| 63 | Rad17 | 12933 | -0.139 | -0.0770 | No |
| 64 | Tcp11 | 13019 | -0.143 | -0.0743 | No |
| 65 | Gapdhs | 13118 | -0.148 | -0.0720 | No |
| 66 | She | 13315 | -0.158 | -0.0743 | No |
| 67 | Bub1 | 13372 | -0.161 | -0.0692 | No |
| 68 | Cdk1 | 13427 | -0.164 | -0.0639 | No |
| 69 | Ybx2 | 13889 | -0.186 | -0.0786 | No |
| 70 | Coil | 14157 | -0.199 | -0.0825 | No |
| 71 | Prkar2a | 14211 | -0.201 | -0.0753 | No |
| 72 | Zc2hc1c | 14285 | -0.205 | -0.0689 | No |
| 73 | Arl4a | 14319 | -0.207 | -0.0603 | No |
| 74 | Ncaph | 14399 | -0.211 | -0.0539 | No |
| 75 | Dmrt1 | 14718 | -0.228 | -0.0590 | No |
| 76 | Nos1 | 14720 | -0.228 | -0.0477 | No |
| 77 | Kif2c | 15196 | -0.251 | -0.0599 | No |
| 78 | Taldo1 | 15243 | -0.254 | -0.0496 | No |
| 79 | Acrbp | 15291 | -0.256 | -0.0393 | No |
| 80 | Mlf1 | 15294 | -0.256 | -0.0266 | No |
| 81 | Tsn | 15312 | -0.257 | -0.0146 | No |
| 82 | Pcsk4 | 15491 | -0.267 | -0.0105 | No |
| 83 | Art3 | 15589 | -0.271 | -0.0021 | No |
| 84 | Parp2 | 15730 | -0.280 | 0.0046 | No |
| 85 | Ttk | 15886 | -0.290 | 0.0110 | No |
| 86 | Dbf4 | 16094 | -0.306 | 0.0155 | No |
| 87 | Phkg2 | 16122 | -0.308 | 0.0295 | No |
| 88 | Camk4 | 16687 | -0.353 | 0.0177 | No |
| 89 | Phf7 | 17142 | -0.391 | 0.0136 | No |
| 90 | Ccnb2 | 17343 | -0.411 | 0.0238 | No |
| 91 | Tulp2 | 17358 | -0.412 | 0.0436 | No |
| 92 | Cdkn3 | 18032 | -0.481 | 0.0326 | No |
| 93 | Pebp1 | 18916 | -0.676 | 0.0203 | No |