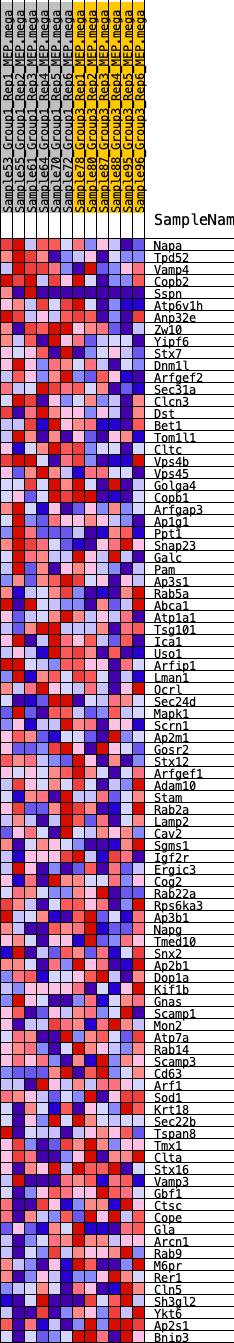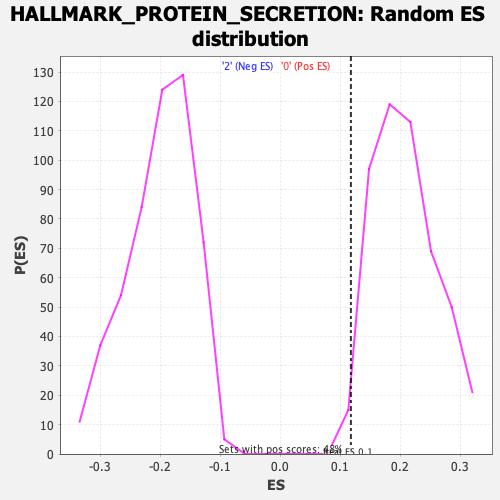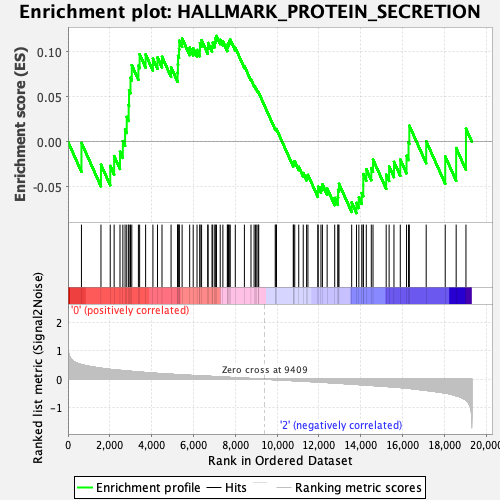
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_PROTEIN_SECRETION |
| Enrichment Score (ES) | 0.11781546 |
| Normalized Enrichment Score (NES) | 0.5683475 |
| Nominal p-value | 0.9855372 |
| FDR q-value | 0.9756168 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Napa | 643 | 0.511 | -0.0012 | Yes |
| 2 | Tpd52 | 1576 | 0.384 | -0.0255 | Yes |
| 3 | Vamp4 | 2019 | 0.342 | -0.0269 | Yes |
| 4 | Copb2 | 2206 | 0.328 | -0.0159 | Yes |
| 5 | Sspn | 2486 | 0.307 | -0.0111 | Yes |
| 6 | Atp6v1h | 2623 | 0.300 | 0.0008 | Yes |
| 7 | Anp32e | 2729 | 0.292 | 0.0138 | Yes |
| 8 | Zw10 | 2809 | 0.287 | 0.0277 | Yes |
| 9 | Yipf6 | 2900 | 0.281 | 0.0408 | Yes |
| 10 | Stx7 | 2919 | 0.279 | 0.0574 | Yes |
| 11 | Dnm1l | 2988 | 0.275 | 0.0713 | Yes |
| 12 | Arfgef2 | 3050 | 0.272 | 0.0852 | Yes |
| 13 | Sec31a | 3366 | 0.252 | 0.0847 | Yes |
| 14 | Clcn3 | 3425 | 0.248 | 0.0973 | Yes |
| 15 | Dst | 3711 | 0.232 | 0.0972 | Yes |
| 16 | Bet1 | 4060 | 0.213 | 0.0925 | Yes |
| 17 | Tom1l1 | 4279 | 0.201 | 0.0938 | Yes |
| 18 | Cltc | 4494 | 0.190 | 0.0947 | Yes |
| 19 | Vps4b | 4928 | 0.171 | 0.0829 | Yes |
| 20 | Vps45 | 5239 | 0.159 | 0.0768 | Yes |
| 21 | Golga4 | 5254 | 0.159 | 0.0861 | Yes |
| 22 | Copb1 | 5263 | 0.158 | 0.0957 | Yes |
| 23 | Arfgap3 | 5309 | 0.157 | 0.1032 | Yes |
| 24 | Ap1g1 | 5321 | 0.156 | 0.1125 | Yes |
| 25 | Ppt1 | 5456 | 0.150 | 0.1150 | Yes |
| 26 | Snap23 | 5817 | 0.134 | 0.1047 | Yes |
| 27 | Galc | 5986 | 0.128 | 0.1041 | Yes |
| 28 | Pam | 6169 | 0.120 | 0.1022 | Yes |
| 29 | Ap3s1 | 6305 | 0.118 | 0.1026 | Yes |
| 30 | Rab5a | 6309 | 0.118 | 0.1099 | Yes |
| 31 | Abca1 | 6387 | 0.114 | 0.1131 | Yes |
| 32 | Atp1a1 | 6682 | 0.103 | 0.1042 | Yes |
| 33 | Tsg101 | 6697 | 0.102 | 0.1100 | Yes |
| 34 | Ica1 | 6886 | 0.095 | 0.1062 | Yes |
| 35 | Uso1 | 6921 | 0.093 | 0.1103 | Yes |
| 36 | Arfip1 | 7022 | 0.089 | 0.1107 | Yes |
| 37 | Lman1 | 7041 | 0.089 | 0.1153 | Yes |
| 38 | Ocrl | 7099 | 0.086 | 0.1178 | Yes |
| 39 | Sec24d | 7278 | 0.080 | 0.1136 | No |
| 40 | Mapk1 | 7405 | 0.075 | 0.1117 | No |
| 41 | Scrn1 | 7617 | 0.066 | 0.1049 | No |
| 42 | Ap2m1 | 7633 | 0.065 | 0.1083 | No |
| 43 | Gosr2 | 7675 | 0.064 | 0.1102 | No |
| 44 | Stx12 | 7710 | 0.063 | 0.1123 | No |
| 45 | Arfgef1 | 7757 | 0.061 | 0.1138 | No |
| 46 | Adam10 | 7999 | 0.053 | 0.1046 | No |
| 47 | Stam | 8434 | 0.035 | 0.0842 | No |
| 48 | Rab2a | 8749 | 0.023 | 0.0693 | No |
| 49 | Lamp2 | 8898 | 0.018 | 0.0627 | No |
| 50 | Cav2 | 8968 | 0.015 | 0.0601 | No |
| 51 | Sgms1 | 9028 | 0.013 | 0.0578 | No |
| 52 | Igf2r | 9092 | 0.011 | 0.0552 | No |
| 53 | Ergic3 | 9119 | 0.010 | 0.0545 | No |
| 54 | Cog2 | 9908 | -0.018 | 0.0146 | No |
| 55 | Rab22a | 9943 | -0.019 | 0.0140 | No |
| 56 | Rps6ka3 | 9968 | -0.020 | 0.0140 | No |
| 57 | Ap3b1 | 10770 | -0.049 | -0.0246 | No |
| 58 | Napg | 10787 | -0.050 | -0.0223 | No |
| 59 | Tmed10 | 10841 | -0.052 | -0.0217 | No |
| 60 | Snx2 | 11032 | -0.059 | -0.0279 | No |
| 61 | Ap2b1 | 11247 | -0.067 | -0.0348 | No |
| 62 | Dop1a | 11407 | -0.074 | -0.0384 | No |
| 63 | Kif1b | 11471 | -0.077 | -0.0368 | No |
| 64 | Gnas | 11947 | -0.096 | -0.0555 | No |
| 65 | Scamp1 | 11959 | -0.097 | -0.0500 | No |
| 66 | Mon2 | 12094 | -0.102 | -0.0505 | No |
| 67 | Atp7a | 12164 | -0.105 | -0.0474 | No |
| 68 | Rab14 | 12390 | -0.115 | -0.0519 | No |
| 69 | Scamp3 | 12752 | -0.130 | -0.0625 | No |
| 70 | Cd63 | 12904 | -0.137 | -0.0617 | No |
| 71 | Arf1 | 12914 | -0.138 | -0.0535 | No |
| 72 | Sod1 | 12956 | -0.140 | -0.0467 | No |
| 73 | Krt18 | 13563 | -0.169 | -0.0676 | No |
| 74 | Sec22b | 13793 | -0.182 | -0.0681 | No |
| 75 | Tspan8 | 13906 | -0.187 | -0.0621 | No |
| 76 | Tmx1 | 14045 | -0.193 | -0.0571 | No |
| 77 | Clta | 14117 | -0.197 | -0.0484 | No |
| 78 | Stx16 | 14118 | -0.197 | -0.0360 | No |
| 79 | Vamp3 | 14262 | -0.204 | -0.0305 | No |
| 80 | Gbf1 | 14501 | -0.215 | -0.0293 | No |
| 81 | Ctsc | 14583 | -0.219 | -0.0197 | No |
| 82 | Cope | 15213 | -0.252 | -0.0366 | No |
| 83 | Gla | 15353 | -0.259 | -0.0274 | No |
| 84 | Arcn1 | 15585 | -0.271 | -0.0224 | No |
| 85 | Rab9 | 15887 | -0.290 | -0.0198 | No |
| 86 | M6pr | 16186 | -0.312 | -0.0156 | No |
| 87 | Rer1 | 16282 | -0.319 | -0.0004 | No |
| 88 | Cln5 | 16320 | -0.321 | 0.0179 | No |
| 89 | Sh3gl2 | 17126 | -0.389 | 0.0006 | No |
| 90 | Ykt6 | 18036 | -0.482 | -0.0164 | No |
| 91 | Ap2s1 | 18559 | -0.579 | -0.0070 | No |
| 92 | Bnip3 | 19027 | -0.728 | 0.0146 | No |