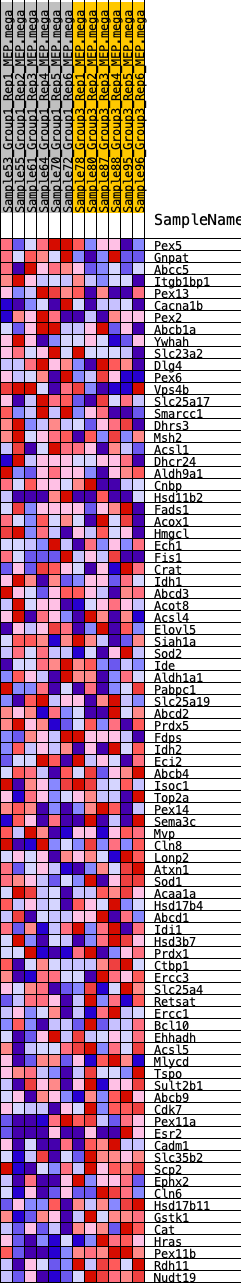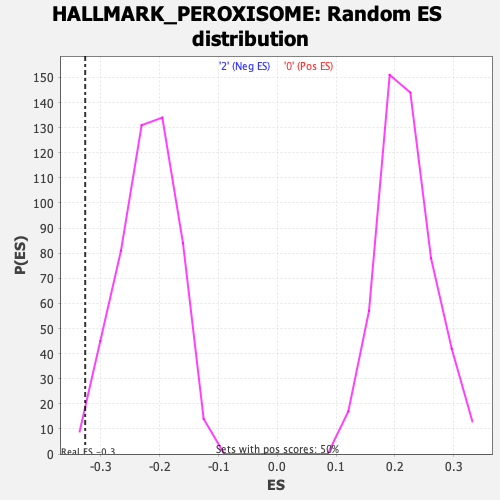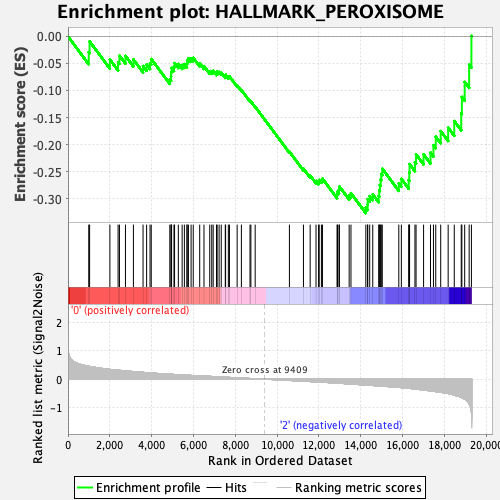
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_PEROXISOME |
| Enrichment Score (ES) | -0.32686788 |
| Normalized Enrichment Score (NES) | -1.485269 |
| Nominal p-value | 0.012048192 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.483 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pex5 | 992 | 0.453 | -0.0292 | No |
| 2 | Gnpat | 1033 | 0.446 | -0.0093 | No |
| 3 | Abcc5 | 2002 | 0.344 | -0.0427 | No |
| 4 | Itgb1bp1 | 2395 | 0.315 | -0.0475 | No |
| 5 | Pex13 | 2460 | 0.310 | -0.0356 | No |
| 6 | Cacna1b | 2747 | 0.291 | -0.0361 | No |
| 7 | Pex2 | 3127 | 0.267 | -0.0426 | No |
| 8 | Abcb1a | 3588 | 0.239 | -0.0547 | No |
| 9 | Ywhah | 3759 | 0.231 | -0.0521 | No |
| 10 | Slc23a2 | 3922 | 0.221 | -0.0496 | No |
| 11 | Dlg4 | 3980 | 0.218 | -0.0418 | No |
| 12 | Pex6 | 4868 | 0.173 | -0.0794 | No |
| 13 | Vps4b | 4928 | 0.171 | -0.0740 | No |
| 14 | Slc25a17 | 4933 | 0.171 | -0.0658 | No |
| 15 | Smarcc1 | 4953 | 0.170 | -0.0584 | No |
| 16 | Dhrs3 | 5067 | 0.166 | -0.0560 | No |
| 17 | Msh2 | 5095 | 0.165 | -0.0493 | No |
| 18 | Acsl1 | 5275 | 0.158 | -0.0508 | No |
| 19 | Dhcr24 | 5457 | 0.150 | -0.0528 | No |
| 20 | Aldh9a1 | 5564 | 0.145 | -0.0512 | No |
| 21 | Cnbp | 5686 | 0.140 | -0.0506 | No |
| 22 | Hsd11b2 | 5697 | 0.139 | -0.0442 | No |
| 23 | Fads1 | 5758 | 0.137 | -0.0406 | No |
| 24 | Acox1 | 5885 | 0.132 | -0.0406 | No |
| 25 | Hmgcl | 5988 | 0.128 | -0.0396 | No |
| 26 | Ech1 | 6297 | 0.118 | -0.0498 | No |
| 27 | Fis1 | 6502 | 0.110 | -0.0550 | No |
| 28 | Crat | 6777 | 0.099 | -0.0643 | No |
| 29 | Idh1 | 6873 | 0.095 | -0.0645 | No |
| 30 | Abcd3 | 6941 | 0.092 | -0.0635 | No |
| 31 | Acot8 | 7100 | 0.086 | -0.0674 | No |
| 32 | Acsl4 | 7131 | 0.086 | -0.0647 | No |
| 33 | Elovl5 | 7225 | 0.082 | -0.0655 | No |
| 34 | Siah1a | 7327 | 0.078 | -0.0669 | No |
| 35 | Sod2 | 7520 | 0.070 | -0.0735 | No |
| 36 | Ide | 7535 | 0.070 | -0.0707 | No |
| 37 | Aldh1a1 | 7667 | 0.064 | -0.0744 | No |
| 38 | Pabpc1 | 7719 | 0.062 | -0.0740 | No |
| 39 | Slc25a19 | 8087 | 0.049 | -0.0906 | No |
| 40 | Abcd2 | 8290 | 0.041 | -0.0991 | No |
| 41 | Prdx5 | 8702 | 0.024 | -0.1193 | No |
| 42 | Fdps | 8744 | 0.023 | -0.1203 | No |
| 43 | Idh2 | 8949 | 0.016 | -0.1301 | No |
| 44 | Eci2 | 10586 | -0.042 | -0.2131 | No |
| 45 | Abcb4 | 11257 | -0.068 | -0.2447 | No |
| 46 | Isoc1 | 11578 | -0.081 | -0.2573 | No |
| 47 | Top2a | 11858 | -0.092 | -0.2673 | No |
| 48 | Pex14 | 11980 | -0.098 | -0.2687 | No |
| 49 | Sema3c | 12010 | -0.099 | -0.2654 | No |
| 50 | Mvp | 12116 | -0.103 | -0.2657 | No |
| 51 | Cln8 | 12166 | -0.105 | -0.2631 | No |
| 52 | Lonp2 | 12864 | -0.136 | -0.2926 | No |
| 53 | Atxn1 | 12885 | -0.136 | -0.2869 | No |
| 54 | Sod1 | 12956 | -0.140 | -0.2836 | No |
| 55 | Acaa1a | 12976 | -0.141 | -0.2776 | No |
| 56 | Hsd17b4 | 13443 | -0.164 | -0.2938 | No |
| 57 | Abcd1 | 13529 | -0.168 | -0.2899 | No |
| 58 | Idi1 | 14240 | -0.203 | -0.3169 | Yes |
| 59 | Hsd3b7 | 14325 | -0.207 | -0.3110 | Yes |
| 60 | Prdx1 | 14329 | -0.207 | -0.3009 | Yes |
| 61 | Ctbp1 | 14425 | -0.212 | -0.2954 | Yes |
| 62 | Ercc3 | 14571 | -0.218 | -0.2922 | Yes |
| 63 | Slc25a4 | 14856 | -0.234 | -0.2953 | Yes |
| 64 | Retsat | 14882 | -0.236 | -0.2850 | Yes |
| 65 | Ercc1 | 14911 | -0.238 | -0.2747 | Yes |
| 66 | Bcl10 | 14947 | -0.240 | -0.2647 | Yes |
| 67 | Ehhadh | 14979 | -0.242 | -0.2543 | Yes |
| 68 | Acsl5 | 15026 | -0.244 | -0.2447 | Yes |
| 69 | Mlycd | 15816 | -0.285 | -0.2717 | Yes |
| 70 | Tspo | 15938 | -0.294 | -0.2634 | Yes |
| 71 | Sult2b1 | 16289 | -0.319 | -0.2659 | Yes |
| 72 | Abcb9 | 16318 | -0.321 | -0.2515 | Yes |
| 73 | Cdk7 | 16331 | -0.322 | -0.2362 | Yes |
| 74 | Pex11a | 16588 | -0.343 | -0.2326 | Yes |
| 75 | Esr2 | 16641 | -0.348 | -0.2181 | Yes |
| 76 | Cadm1 | 17001 | -0.379 | -0.2180 | Yes |
| 77 | Slc35b2 | 17331 | -0.410 | -0.2149 | Yes |
| 78 | Scp2 | 17473 | -0.425 | -0.2013 | Yes |
| 79 | Ephx2 | 17583 | -0.433 | -0.1855 | Yes |
| 80 | Cln6 | 17820 | -0.460 | -0.1751 | Yes |
| 81 | Hsd17b11 | 18176 | -0.501 | -0.1687 | Yes |
| 82 | Gstk1 | 18468 | -0.555 | -0.1565 | Yes |
| 83 | Cat | 18801 | -0.637 | -0.1423 | Yes |
| 84 | Hras | 18832 | -0.646 | -0.1119 | Yes |
| 85 | Pex11b | 18966 | -0.701 | -0.0842 | Yes |
| 86 | Rdh11 | 19180 | -0.871 | -0.0522 | Yes |
| 87 | Nudt19 | 19288 | -1.190 | 0.0010 | Yes |