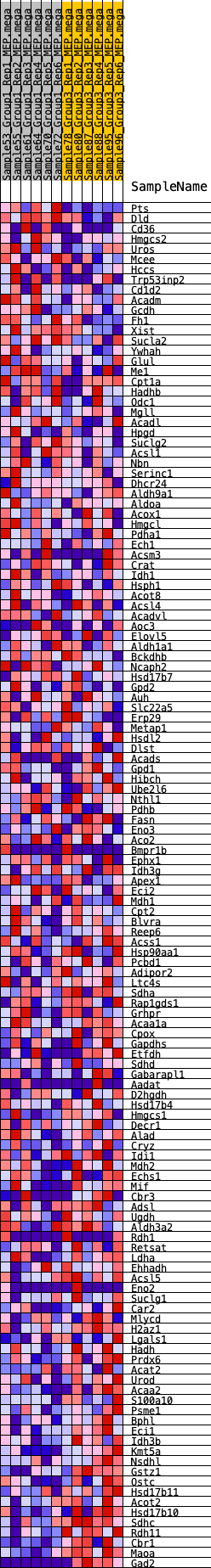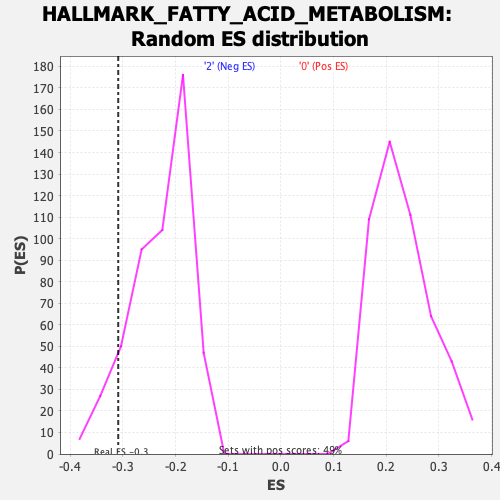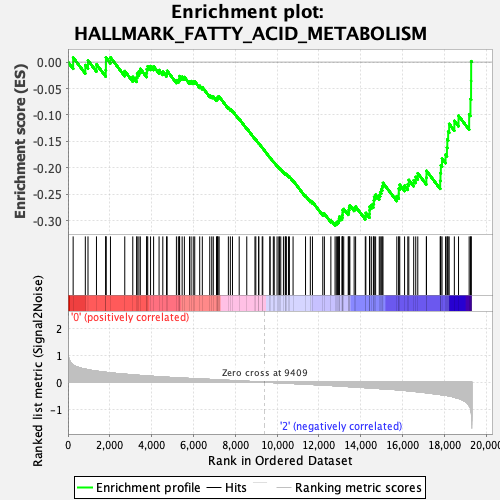
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_FATTY_ACID_METABOLISM |
| Enrichment Score (ES) | -0.3090741 |
| Normalized Enrichment Score (NES) | -1.3527552 |
| Nominal p-value | 0.096837945 |
| FDR q-value | 0.7435843 |
| FWER p-Value | 0.739 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pts | 246 | 0.638 | 0.0085 | No |
| 2 | Dld | 828 | 0.480 | -0.0057 | No |
| 3 | Cd36 | 956 | 0.459 | 0.0031 | No |
| 4 | Hmgcs2 | 1361 | 0.406 | -0.0044 | No |
| 5 | Uros | 1805 | 0.363 | -0.0153 | No |
| 6 | Mcee | 1813 | 0.362 | -0.0036 | No |
| 7 | Hccs | 1814 | 0.362 | 0.0086 | No |
| 8 | Trp53inp2 | 2030 | 0.341 | 0.0088 | No |
| 9 | Cd1d2 | 2714 | 0.294 | -0.0170 | No |
| 10 | Acadm | 3097 | 0.269 | -0.0279 | No |
| 11 | Gcdh | 3281 | 0.256 | -0.0289 | No |
| 12 | Fh1 | 3306 | 0.255 | -0.0216 | No |
| 13 | Xist | 3390 | 0.250 | -0.0175 | No |
| 14 | Sucla2 | 3458 | 0.246 | -0.0128 | No |
| 15 | Ywhah | 3759 | 0.231 | -0.0207 | No |
| 16 | Glul | 3769 | 0.231 | -0.0134 | No |
| 17 | Me1 | 3812 | 0.228 | -0.0080 | No |
| 18 | Cpt1a | 3947 | 0.220 | -0.0076 | No |
| 19 | Hadhb | 4093 | 0.211 | -0.0081 | No |
| 20 | Odc1 | 4354 | 0.197 | -0.0150 | No |
| 21 | Mgll | 4533 | 0.188 | -0.0180 | No |
| 22 | Acadl | 4711 | 0.181 | -0.0212 | No |
| 23 | Hpgd | 4739 | 0.179 | -0.0166 | No |
| 24 | Suclg2 | 5183 | 0.161 | -0.0343 | No |
| 25 | Acsl1 | 5275 | 0.158 | -0.0338 | No |
| 26 | Nbn | 5326 | 0.156 | -0.0312 | No |
| 27 | Serinc1 | 5337 | 0.155 | -0.0265 | No |
| 28 | Dhcr24 | 5457 | 0.150 | -0.0277 | No |
| 29 | Aldh9a1 | 5564 | 0.145 | -0.0283 | No |
| 30 | Aldoa | 5809 | 0.135 | -0.0365 | No |
| 31 | Acox1 | 5885 | 0.132 | -0.0360 | No |
| 32 | Hmgcl | 5988 | 0.128 | -0.0371 | No |
| 33 | Pdha1 | 6059 | 0.125 | -0.0365 | No |
| 34 | Ech1 | 6297 | 0.118 | -0.0449 | No |
| 35 | Acsm3 | 6422 | 0.113 | -0.0476 | No |
| 36 | Crat | 6777 | 0.099 | -0.0628 | No |
| 37 | Idh1 | 6873 | 0.095 | -0.0645 | No |
| 38 | Hsph1 | 6946 | 0.092 | -0.0652 | No |
| 39 | Acot8 | 7100 | 0.086 | -0.0703 | No |
| 40 | Acsl4 | 7131 | 0.086 | -0.0690 | No |
| 41 | Acadvl | 7151 | 0.085 | -0.0671 | No |
| 42 | Aoc3 | 7174 | 0.084 | -0.0654 | No |
| 43 | Elovl5 | 7225 | 0.082 | -0.0653 | No |
| 44 | Aldh1a1 | 7667 | 0.064 | -0.0861 | No |
| 45 | Bckdhb | 7762 | 0.061 | -0.0890 | No |
| 46 | Ncaph2 | 7864 | 0.057 | -0.0924 | No |
| 47 | Hsd17b7 | 8185 | 0.045 | -0.1075 | No |
| 48 | Gpd2 | 8547 | 0.031 | -0.1253 | No |
| 49 | Auh | 8939 | 0.016 | -0.1452 | No |
| 50 | Slc22a5 | 8980 | 0.015 | -0.1468 | No |
| 51 | Erp29 | 9115 | 0.010 | -0.1534 | No |
| 52 | Metap1 | 9122 | 0.010 | -0.1534 | No |
| 53 | Hsdl2 | 9286 | 0.004 | -0.1618 | No |
| 54 | Dlst | 9314 | 0.003 | -0.1631 | No |
| 55 | Acads | 9653 | -0.007 | -0.1805 | No |
| 56 | Gpd1 | 9660 | -0.007 | -0.1805 | No |
| 57 | Hibch | 9817 | -0.014 | -0.1882 | No |
| 58 | Ube2l6 | 9860 | -0.015 | -0.1899 | No |
| 59 | Nthl1 | 9996 | -0.021 | -0.1962 | No |
| 60 | Pdhb | 10079 | -0.024 | -0.1997 | No |
| 61 | Fasn | 10116 | -0.026 | -0.2007 | No |
| 62 | Eno3 | 10173 | -0.028 | -0.2027 | No |
| 63 | Aco2 | 10300 | -0.031 | -0.2082 | No |
| 64 | Bmpr1b | 10388 | -0.034 | -0.2116 | No |
| 65 | Ephx1 | 10396 | -0.034 | -0.2108 | No |
| 66 | Idh3g | 10441 | -0.036 | -0.2119 | No |
| 67 | Apex1 | 10541 | -0.041 | -0.2157 | No |
| 68 | Eci2 | 10586 | -0.042 | -0.2166 | No |
| 69 | Mdh1 | 10761 | -0.049 | -0.2240 | No |
| 70 | Cpt2 | 11357 | -0.072 | -0.2527 | No |
| 71 | Blvra | 11586 | -0.081 | -0.2618 | No |
| 72 | Reep6 | 11688 | -0.086 | -0.2642 | No |
| 73 | Acss1 | 12181 | -0.106 | -0.2863 | No |
| 74 | Hsp90aa1 | 12258 | -0.109 | -0.2866 | No |
| 75 | Pcbd1 | 12568 | -0.122 | -0.2986 | No |
| 76 | Adipor2 | 12769 | -0.131 | -0.3047 | Yes |
| 77 | Ltc4s | 12829 | -0.134 | -0.3033 | Yes |
| 78 | Sdha | 12883 | -0.136 | -0.3015 | Yes |
| 79 | Rap1gds1 | 12937 | -0.139 | -0.2995 | Yes |
| 80 | Grhpr | 12962 | -0.141 | -0.2961 | Yes |
| 81 | Acaa1a | 12976 | -0.141 | -0.2920 | Yes |
| 82 | Cpox | 13099 | -0.147 | -0.2935 | Yes |
| 83 | Gapdhs | 13118 | -0.148 | -0.2895 | Yes |
| 84 | Etfdh | 13128 | -0.148 | -0.2850 | Yes |
| 85 | Sdhd | 13129 | -0.148 | -0.2800 | Yes |
| 86 | Gabarapl1 | 13183 | -0.151 | -0.2777 | Yes |
| 87 | Aadat | 13398 | -0.162 | -0.2834 | Yes |
| 88 | D2hgdh | 13428 | -0.164 | -0.2795 | Yes |
| 89 | Hsd17b4 | 13443 | -0.164 | -0.2747 | Yes |
| 90 | Hmgcs1 | 13476 | -0.165 | -0.2708 | Yes |
| 91 | Decr1 | 13681 | -0.175 | -0.2756 | Yes |
| 92 | Alad | 13751 | -0.179 | -0.2732 | Yes |
| 93 | Cryz | 14214 | -0.201 | -0.2906 | Yes |
| 94 | Idi1 | 14240 | -0.203 | -0.2851 | Yes |
| 95 | Mdh2 | 14415 | -0.211 | -0.2871 | Yes |
| 96 | Echs1 | 14419 | -0.212 | -0.2801 | Yes |
| 97 | Mif | 14424 | -0.212 | -0.2732 | Yes |
| 98 | Cbr3 | 14503 | -0.215 | -0.2701 | Yes |
| 99 | Adsl | 14600 | -0.220 | -0.2677 | Yes |
| 100 | Ugdh | 14624 | -0.222 | -0.2615 | Yes |
| 101 | Aldh3a2 | 14637 | -0.223 | -0.2546 | Yes |
| 102 | Rdh1 | 14708 | -0.228 | -0.2507 | Yes |
| 103 | Retsat | 14882 | -0.236 | -0.2518 | Yes |
| 104 | Ldha | 14926 | -0.238 | -0.2460 | Yes |
| 105 | Ehhadh | 14979 | -0.242 | -0.2406 | Yes |
| 106 | Acsl5 | 15026 | -0.244 | -0.2349 | Yes |
| 107 | Eno2 | 15060 | -0.246 | -0.2284 | Yes |
| 108 | Suclg1 | 15721 | -0.279 | -0.2534 | Yes |
| 109 | Car2 | 15815 | -0.285 | -0.2487 | Yes |
| 110 | Mlycd | 15816 | -0.285 | -0.2392 | Yes |
| 111 | H2az1 | 15862 | -0.288 | -0.2319 | Yes |
| 112 | Lgals1 | 16092 | -0.306 | -0.2336 | Yes |
| 113 | Hadh | 16247 | -0.316 | -0.2310 | Yes |
| 114 | Prdx6 | 16295 | -0.319 | -0.2228 | Yes |
| 115 | Acat2 | 16528 | -0.338 | -0.2235 | Yes |
| 116 | Urod | 16626 | -0.346 | -0.2170 | Yes |
| 117 | Acaa2 | 16724 | -0.355 | -0.2102 | Yes |
| 118 | S100a10 | 17133 | -0.390 | -0.2184 | Yes |
| 119 | Psme1 | 17140 | -0.391 | -0.2056 | Yes |
| 120 | Bphl | 17795 | -0.456 | -0.2244 | Yes |
| 121 | Eci1 | 17810 | -0.459 | -0.2097 | Yes |
| 122 | Idh3b | 17828 | -0.461 | -0.1952 | Yes |
| 123 | Kmt5a | 17883 | -0.466 | -0.1824 | Yes |
| 124 | Nsdhl | 18051 | -0.482 | -0.1749 | Yes |
| 125 | Gstz1 | 18118 | -0.493 | -0.1619 | Yes |
| 126 | Ostc | 18136 | -0.495 | -0.1462 | Yes |
| 127 | Hsd17b11 | 18176 | -0.501 | -0.1314 | Yes |
| 128 | Acot2 | 18224 | -0.508 | -0.1168 | Yes |
| 129 | Hsd17b10 | 18472 | -0.555 | -0.1111 | Yes |
| 130 | Sdhc | 18672 | -0.599 | -0.1014 | Yes |
| 131 | Rdh11 | 19180 | -0.871 | -0.0986 | Yes |
| 132 | Cbr1 | 19242 | -0.952 | -0.0699 | Yes |
| 133 | Maoa | 19272 | -1.070 | -0.0356 | Yes |
| 134 | Gad2 | 19276 | -1.115 | 0.0016 | Yes |