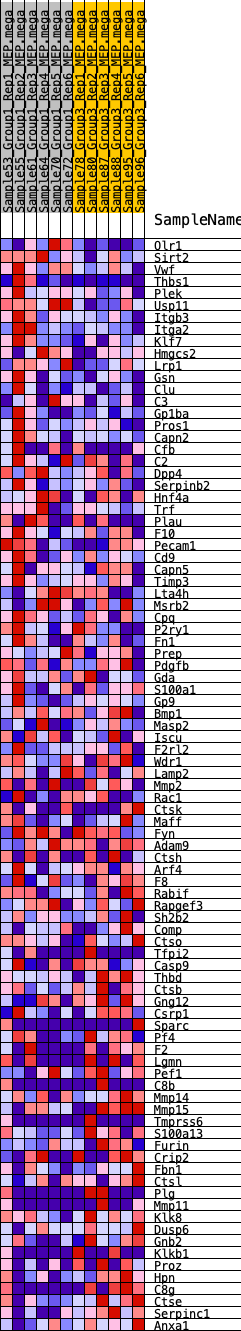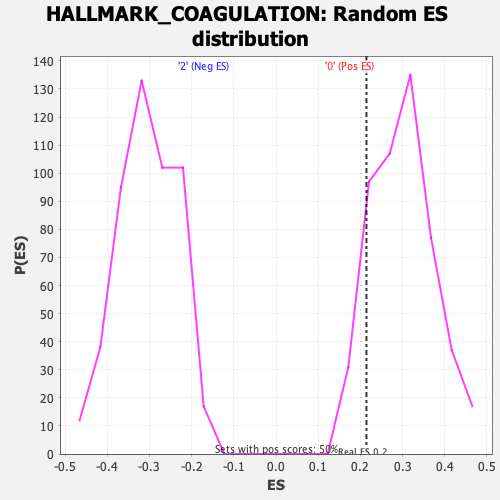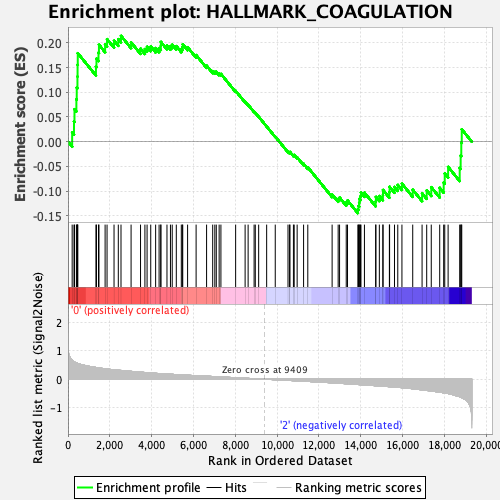
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_COAGULATION |
| Enrichment Score (ES) | 0.21436575 |
| Normalized Enrichment Score (NES) | 0.71225154 |
| Nominal p-value | 0.8742515 |
| FDR q-value | 0.9650142 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Olr1 | 199 | 0.671 | 0.0188 | Yes |
| 2 | Sirt2 | 287 | 0.612 | 0.0408 | Yes |
| 3 | Vwf | 310 | 0.600 | 0.0658 | Yes |
| 4 | Thbs1 | 401 | 0.570 | 0.0859 | Yes |
| 5 | Plek | 418 | 0.565 | 0.1096 | Yes |
| 6 | Usp11 | 452 | 0.555 | 0.1320 | Yes |
| 7 | Itgb3 | 458 | 0.552 | 0.1557 | Yes |
| 8 | Itga2 | 467 | 0.551 | 0.1792 | Yes |
| 9 | Klf7 | 1336 | 0.409 | 0.1518 | Yes |
| 10 | Hmgcs2 | 1361 | 0.406 | 0.1682 | Yes |
| 11 | Lrp1 | 1458 | 0.396 | 0.1805 | Yes |
| 12 | Gsn | 1480 | 0.393 | 0.1964 | Yes |
| 13 | Clu | 1774 | 0.365 | 0.1970 | Yes |
| 14 | C3 | 1867 | 0.356 | 0.2077 | Yes |
| 15 | Gp1ba | 2201 | 0.328 | 0.2046 | Yes |
| 16 | Pros1 | 2405 | 0.314 | 0.2077 | Yes |
| 17 | Capn2 | 2534 | 0.307 | 0.2144 | Yes |
| 18 | Cfb | 3016 | 0.274 | 0.2012 | No |
| 19 | C2 | 3468 | 0.245 | 0.1884 | No |
| 20 | Dpp4 | 3677 | 0.234 | 0.1878 | No |
| 21 | Serpinb2 | 3781 | 0.230 | 0.1924 | No |
| 22 | Hnf4a | 3956 | 0.219 | 0.1929 | No |
| 23 | Trf | 4190 | 0.206 | 0.1897 | No |
| 24 | Plau | 4355 | 0.197 | 0.1897 | No |
| 25 | F10 | 4437 | 0.193 | 0.1939 | No |
| 26 | Pecam1 | 4440 | 0.193 | 0.2022 | No |
| 27 | Cd9 | 4732 | 0.180 | 0.1948 | No |
| 28 | Capn5 | 4898 | 0.172 | 0.1937 | No |
| 29 | Timp3 | 4983 | 0.169 | 0.1967 | No |
| 30 | Lta4h | 5178 | 0.161 | 0.1936 | No |
| 31 | Msrb2 | 5417 | 0.152 | 0.1878 | No |
| 32 | Cpq | 5467 | 0.150 | 0.1917 | No |
| 33 | P2ry1 | 5486 | 0.149 | 0.1973 | No |
| 34 | Fn1 | 5718 | 0.138 | 0.1913 | No |
| 35 | Prep | 6125 | 0.123 | 0.1755 | No |
| 36 | Pdgfb | 6626 | 0.105 | 0.1540 | No |
| 37 | Gda | 6918 | 0.093 | 0.1429 | No |
| 38 | S100a1 | 7020 | 0.089 | 0.1415 | No |
| 39 | Gp9 | 7090 | 0.087 | 0.1417 | No |
| 40 | Bmp1 | 7226 | 0.082 | 0.1382 | No |
| 41 | Masp2 | 7311 | 0.079 | 0.1373 | No |
| 42 | Iscu | 8015 | 0.052 | 0.1030 | No |
| 43 | F2rl2 | 8468 | 0.034 | 0.0809 | No |
| 44 | Wdr1 | 8612 | 0.029 | 0.0747 | No |
| 45 | Lamp2 | 8898 | 0.018 | 0.0607 | No |
| 46 | Mmp2 | 8955 | 0.016 | 0.0584 | No |
| 47 | Rac1 | 9116 | 0.010 | 0.0505 | No |
| 48 | Ctsk | 9498 | -0.001 | 0.0307 | No |
| 49 | Maff | 9909 | -0.018 | 0.0102 | No |
| 50 | Fyn | 10519 | -0.040 | -0.0198 | No |
| 51 | Adam9 | 10595 | -0.042 | -0.0218 | No |
| 52 | Ctsh | 10611 | -0.043 | -0.0208 | No |
| 53 | Arf4 | 10778 | -0.049 | -0.0273 | No |
| 54 | F8 | 10814 | -0.051 | -0.0269 | No |
| 55 | Rabif | 10956 | -0.057 | -0.0318 | No |
| 56 | Rapgef3 | 11264 | -0.068 | -0.0448 | No |
| 57 | Sh2b2 | 11465 | -0.077 | -0.0519 | No |
| 58 | Comp | 12628 | -0.125 | -0.1069 | No |
| 59 | Ctso | 12917 | -0.138 | -0.1159 | No |
| 60 | Tfpi2 | 12984 | -0.141 | -0.1132 | No |
| 61 | Casp9 | 13303 | -0.157 | -0.1229 | No |
| 62 | Thbd | 13364 | -0.160 | -0.1191 | No |
| 63 | Ctsb | 13859 | -0.185 | -0.1368 | No |
| 64 | Gng12 | 13890 | -0.186 | -0.1302 | No |
| 65 | Csrp1 | 13923 | -0.188 | -0.1237 | No |
| 66 | Sparc | 13929 | -0.188 | -0.1158 | No |
| 67 | Pf4 | 13986 | -0.189 | -0.1105 | No |
| 68 | F2 | 14006 | -0.191 | -0.1032 | No |
| 69 | Lgmn | 14173 | -0.199 | -0.1032 | No |
| 70 | Pef1 | 14710 | -0.228 | -0.1212 | No |
| 71 | C8b | 14721 | -0.228 | -0.1118 | No |
| 72 | Mmp14 | 14886 | -0.236 | -0.1101 | No |
| 73 | Mmp15 | 15056 | -0.245 | -0.1083 | No |
| 74 | Tmprss6 | 15061 | -0.246 | -0.0978 | No |
| 75 | S100a13 | 15366 | -0.260 | -0.1023 | No |
| 76 | Furin | 15376 | -0.261 | -0.0914 | No |
| 77 | Crip2 | 15610 | -0.273 | -0.0917 | No |
| 78 | Fbn1 | 15767 | -0.282 | -0.0876 | No |
| 79 | Ctsl | 15966 | -0.295 | -0.0851 | No |
| 80 | Plg | 16482 | -0.334 | -0.0974 | No |
| 81 | Mmp11 | 16929 | -0.372 | -0.1044 | No |
| 82 | Klk8 | 17150 | -0.392 | -0.0988 | No |
| 83 | Dusp6 | 17368 | -0.413 | -0.0922 | No |
| 84 | Gnb2 | 17772 | -0.453 | -0.0935 | No |
| 85 | Klkb1 | 17962 | -0.474 | -0.0827 | No |
| 86 | Proz | 18013 | -0.479 | -0.0645 | No |
| 87 | Hpn | 18174 | -0.501 | -0.0511 | No |
| 88 | C8g | 18728 | -0.616 | -0.0531 | No |
| 89 | Ctse | 18774 | -0.631 | -0.0281 | No |
| 90 | Serpinc1 | 18804 | -0.638 | -0.0019 | No |
| 91 | Anxa1 | 18826 | -0.644 | 0.0250 | No |