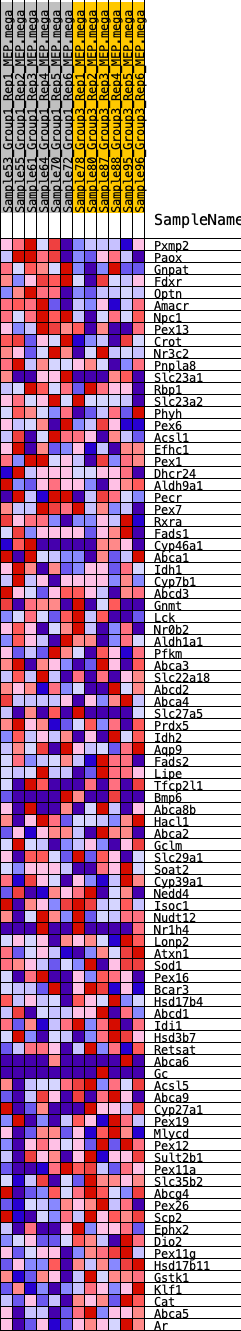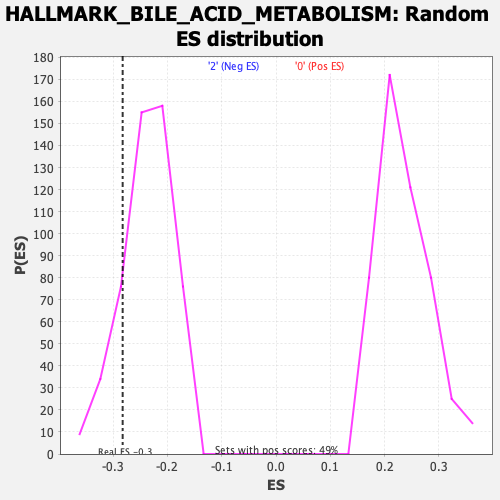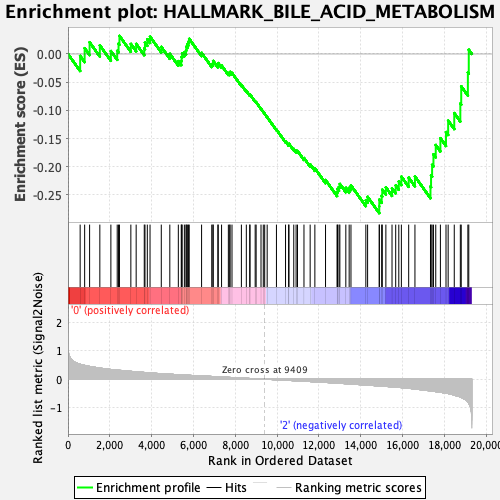
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group3.MEP.mega_Pheno.cls #Group1_versus_Group3_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group3_repos |
| Upregulated in class | 2 |
| GeneSet | HALLMARK_BILE_ACID_METABOLISM |
| Enrichment Score (ES) | -0.28238106 |
| Normalized Enrichment Score (NES) | -1.19132 |
| Nominal p-value | 0.1437008 |
| FDR q-value | 0.77260345 |
| FWER p-Value | 0.935 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Pxmp2 | 581 | 0.524 | -0.0034 | No |
| 2 | Paox | 796 | 0.483 | 0.0103 | No |
| 3 | Gnpat | 1033 | 0.446 | 0.0208 | No |
| 4 | Fdxr | 1522 | 0.388 | 0.0153 | No |
| 5 | Optn | 2050 | 0.339 | 0.0052 | No |
| 6 | Amacr | 2353 | 0.317 | 0.0058 | No |
| 7 | Npc1 | 2417 | 0.312 | 0.0185 | No |
| 8 | Pex13 | 2460 | 0.310 | 0.0321 | No |
| 9 | Crot | 3005 | 0.274 | 0.0179 | No |
| 10 | Nr3c2 | 3259 | 0.258 | 0.0179 | No |
| 11 | Pnpla8 | 3646 | 0.236 | 0.0099 | No |
| 12 | Slc23a1 | 3680 | 0.234 | 0.0202 | No |
| 13 | Rbp1 | 3792 | 0.229 | 0.0261 | No |
| 14 | Slc23a2 | 3922 | 0.221 | 0.0308 | No |
| 15 | Phyh | 4460 | 0.192 | 0.0127 | No |
| 16 | Pex6 | 4868 | 0.173 | 0.0004 | No |
| 17 | Acsl1 | 5275 | 0.158 | -0.0127 | No |
| 18 | Efhc1 | 5414 | 0.152 | -0.0121 | No |
| 19 | Pex1 | 5433 | 0.151 | -0.0053 | No |
| 20 | Dhcr24 | 5457 | 0.150 | 0.0012 | No |
| 21 | Aldh9a1 | 5564 | 0.145 | 0.0031 | No |
| 22 | Pecr | 5644 | 0.142 | 0.0063 | No |
| 23 | Pex7 | 5655 | 0.142 | 0.0130 | No |
| 24 | Rxra | 5707 | 0.139 | 0.0175 | No |
| 25 | Fads1 | 5758 | 0.137 | 0.0219 | No |
| 26 | Cyp46a1 | 5795 | 0.135 | 0.0269 | No |
| 27 | Abca1 | 6387 | 0.114 | 0.0020 | No |
| 28 | Idh1 | 6873 | 0.095 | -0.0183 | No |
| 29 | Cyp7b1 | 6930 | 0.093 | -0.0165 | No |
| 30 | Abcd3 | 6941 | 0.092 | -0.0123 | No |
| 31 | Gnmt | 7166 | 0.084 | -0.0196 | No |
| 32 | Lck | 7182 | 0.084 | -0.0161 | No |
| 33 | Nr0b2 | 7342 | 0.077 | -0.0204 | No |
| 34 | Aldh1a1 | 7667 | 0.064 | -0.0340 | No |
| 35 | Pfkm | 7735 | 0.062 | -0.0343 | No |
| 36 | Abca3 | 7746 | 0.061 | -0.0317 | No |
| 37 | Slc22a18 | 7841 | 0.058 | -0.0336 | No |
| 38 | Abcd2 | 8290 | 0.041 | -0.0548 | No |
| 39 | Abca4 | 8526 | 0.032 | -0.0654 | No |
| 40 | Slc27a5 | 8689 | 0.025 | -0.0726 | No |
| 41 | Prdx5 | 8702 | 0.024 | -0.0719 | No |
| 42 | Idh2 | 8949 | 0.016 | -0.0839 | No |
| 43 | Aqp9 | 8995 | 0.014 | -0.0855 | No |
| 44 | Fads2 | 9228 | 0.006 | -0.0973 | No |
| 45 | Lipe | 9337 | 0.003 | -0.1028 | No |
| 46 | Tfcp2l1 | 9404 | 0.000 | -0.1062 | No |
| 47 | Bmp6 | 9523 | -0.002 | -0.1122 | No |
| 48 | Abca8b | 9966 | -0.020 | -0.1342 | No |
| 49 | Hacl1 | 10401 | -0.035 | -0.1550 | No |
| 50 | Abca2 | 10549 | -0.041 | -0.1606 | No |
| 51 | Gclm | 10564 | -0.041 | -0.1592 | No |
| 52 | Slc29a1 | 10794 | -0.050 | -0.1685 | No |
| 53 | Soat2 | 10906 | -0.055 | -0.1715 | No |
| 54 | Cyp39a1 | 10972 | -0.057 | -0.1720 | No |
| 55 | Nedd4 | 11282 | -0.068 | -0.1846 | No |
| 56 | Isoc1 | 11578 | -0.081 | -0.1957 | No |
| 57 | Nudt12 | 11803 | -0.090 | -0.2028 | No |
| 58 | Nr1h4 | 12313 | -0.111 | -0.2236 | No |
| 59 | Lonp2 | 12864 | -0.136 | -0.2452 | No |
| 60 | Atxn1 | 12885 | -0.136 | -0.2393 | No |
| 61 | Sod1 | 12956 | -0.140 | -0.2357 | No |
| 62 | Pex16 | 12998 | -0.142 | -0.2306 | No |
| 63 | Bcar3 | 13283 | -0.156 | -0.2374 | No |
| 64 | Hsd17b4 | 13443 | -0.164 | -0.2373 | No |
| 65 | Abcd1 | 13529 | -0.168 | -0.2331 | No |
| 66 | Idi1 | 14240 | -0.203 | -0.2597 | No |
| 67 | Hsd3b7 | 14325 | -0.207 | -0.2534 | No |
| 68 | Retsat | 14882 | -0.236 | -0.2703 | Yes |
| 69 | Abca6 | 14885 | -0.236 | -0.2583 | Yes |
| 70 | Gc | 14998 | -0.243 | -0.2517 | Yes |
| 71 | Acsl5 | 15026 | -0.244 | -0.2406 | Yes |
| 72 | Abca9 | 15198 | -0.251 | -0.2367 | Yes |
| 73 | Cyp27a1 | 15492 | -0.267 | -0.2382 | Yes |
| 74 | Pex19 | 15668 | -0.276 | -0.2332 | Yes |
| 75 | Mlycd | 15816 | -0.285 | -0.2263 | Yes |
| 76 | Pex12 | 15939 | -0.294 | -0.2176 | Yes |
| 77 | Sult2b1 | 16289 | -0.319 | -0.2194 | Yes |
| 78 | Pex11a | 16588 | -0.343 | -0.2173 | Yes |
| 79 | Slc35b2 | 17331 | -0.410 | -0.2350 | Yes |
| 80 | Abcg4 | 17359 | -0.412 | -0.2153 | Yes |
| 81 | Pex26 | 17409 | -0.417 | -0.1964 | Yes |
| 82 | Scp2 | 17473 | -0.425 | -0.1780 | Yes |
| 83 | Ephx2 | 17583 | -0.433 | -0.1614 | Yes |
| 84 | Dio2 | 17805 | -0.459 | -0.1494 | Yes |
| 85 | Pex11g | 18073 | -0.485 | -0.1385 | Yes |
| 86 | Hsd17b11 | 18176 | -0.501 | -0.1181 | Yes |
| 87 | Gstk1 | 18468 | -0.555 | -0.1048 | Yes |
| 88 | Klf1 | 18758 | -0.626 | -0.0878 | Yes |
| 89 | Cat | 18801 | -0.637 | -0.0573 | Yes |
| 90 | Abca5 | 19119 | -0.797 | -0.0330 | Yes |
| 91 | Ar | 19162 | -0.835 | 0.0075 | Yes |