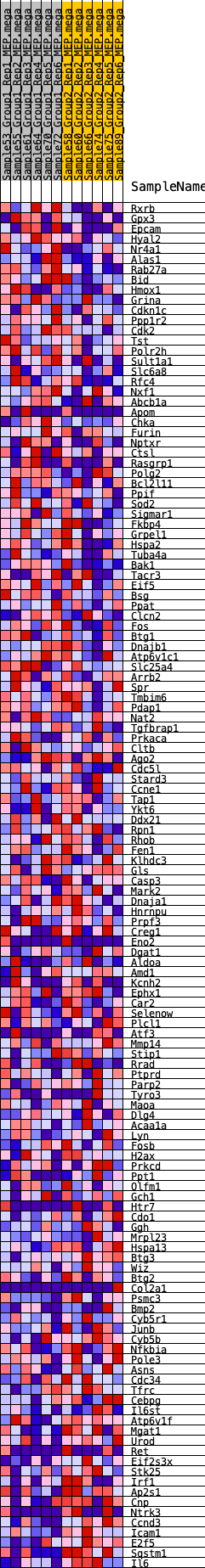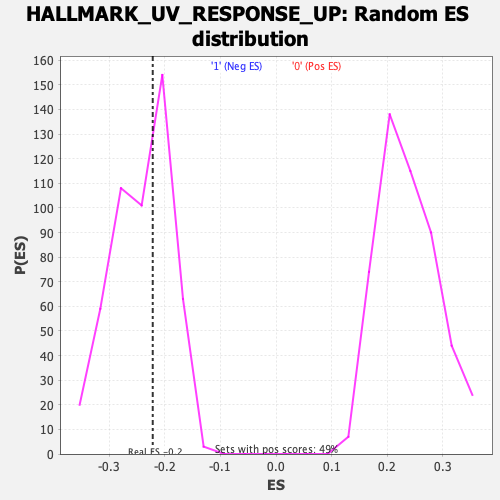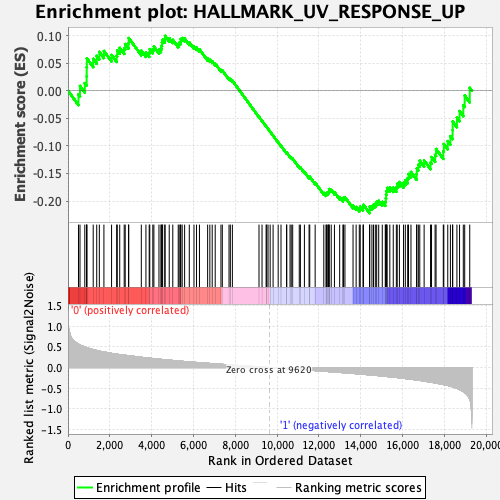
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 1 |
| GeneSet | HALLMARK_UV_RESPONSE_UP |
| Enrichment Score (ES) | -0.22197968 |
| Normalized Enrichment Score (NES) | -0.92019135 |
| Nominal p-value | 0.5748032 |
| FDR q-value | 0.94993335 |
| FWER p-Value | 0.991 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Rxrb | 501 | 0.565 | -0.0066 | No |
| 2 | Gpx3 | 573 | 0.549 | 0.0086 | No |
| 3 | Epcam | 803 | 0.497 | 0.0138 | No |
| 4 | Hyal2 | 891 | 0.481 | 0.0258 | No |
| 5 | Nr4a1 | 894 | 0.480 | 0.0423 | No |
| 6 | Alas1 | 901 | 0.479 | 0.0585 | No |
| 7 | Rab27a | 1205 | 0.432 | 0.0576 | No |
| 8 | Bid | 1373 | 0.411 | 0.0631 | No |
| 9 | Hmox1 | 1498 | 0.398 | 0.0704 | No |
| 10 | Grina | 1717 | 0.377 | 0.0720 | No |
| 11 | Cdkn1c | 2083 | 0.342 | 0.0647 | No |
| 12 | Ppp1r2 | 2327 | 0.324 | 0.0633 | No |
| 13 | Cdk2 | 2354 | 0.322 | 0.0730 | No |
| 14 | Tst | 2473 | 0.312 | 0.0776 | No |
| 15 | Polr2h | 2687 | 0.299 | 0.0768 | No |
| 16 | Sult1a1 | 2735 | 0.295 | 0.0845 | No |
| 17 | Slc6a8 | 2895 | 0.285 | 0.0861 | No |
| 18 | Rfc4 | 2904 | 0.284 | 0.0954 | No |
| 19 | Nxf1 | 3505 | 0.247 | 0.0727 | No |
| 20 | Abcb1a | 3726 | 0.233 | 0.0692 | No |
| 21 | Apom | 3882 | 0.226 | 0.0689 | No |
| 22 | Chka | 3910 | 0.225 | 0.0753 | No |
| 23 | Furin | 4055 | 0.216 | 0.0753 | No |
| 24 | Nptxr | 4106 | 0.213 | 0.0800 | No |
| 25 | Ctsl | 4349 | 0.202 | 0.0743 | No |
| 26 | Rasgrp1 | 4442 | 0.197 | 0.0763 | No |
| 27 | Polg2 | 4478 | 0.195 | 0.0813 | No |
| 28 | Bcl2l11 | 4493 | 0.195 | 0.0872 | No |
| 29 | Ppif | 4524 | 0.193 | 0.0923 | No |
| 30 | Sod2 | 4628 | 0.188 | 0.0935 | No |
| 31 | Sigmar1 | 4640 | 0.187 | 0.0994 | No |
| 32 | Fkbp4 | 4840 | 0.178 | 0.0951 | No |
| 33 | Grpel1 | 5010 | 0.170 | 0.0922 | No |
| 34 | Hspa2 | 5270 | 0.158 | 0.0841 | No |
| 35 | Tuba4a | 5316 | 0.155 | 0.0871 | No |
| 36 | Bak1 | 5382 | 0.153 | 0.0890 | No |
| 37 | Tacr3 | 5392 | 0.152 | 0.0938 | No |
| 38 | Eif5 | 5459 | 0.150 | 0.0955 | No |
| 39 | Bsg | 5576 | 0.145 | 0.0944 | No |
| 40 | Ppat | 5799 | 0.136 | 0.0875 | No |
| 41 | Clcn2 | 6023 | 0.126 | 0.0802 | No |
| 42 | Fos | 6145 | 0.122 | 0.0781 | No |
| 43 | Btg1 | 6285 | 0.117 | 0.0749 | No |
| 44 | Dnajb1 | 6676 | 0.102 | 0.0581 | No |
| 45 | Atp6v1c1 | 6782 | 0.097 | 0.0560 | No |
| 46 | Slc25a4 | 6895 | 0.094 | 0.0534 | No |
| 47 | Arrb2 | 7038 | 0.088 | 0.0490 | No |
| 48 | Spr | 7313 | 0.078 | 0.0374 | No |
| 49 | Tmbim6 | 7378 | 0.075 | 0.0367 | No |
| 50 | Pdap1 | 7699 | 0.065 | 0.0223 | No |
| 51 | Nat2 | 7761 | 0.063 | 0.0212 | No |
| 52 | Tgfbrap1 | 7859 | 0.059 | 0.0182 | No |
| 53 | Prkaca | 9132 | 0.016 | -0.0476 | No |
| 54 | Cltb | 9277 | 0.011 | -0.0547 | No |
| 55 | Ago2 | 9472 | 0.005 | -0.0646 | No |
| 56 | Cdc5l | 9500 | 0.004 | -0.0659 | No |
| 57 | Stard3 | 9564 | 0.002 | -0.0691 | No |
| 58 | Ccne1 | 9666 | -0.000 | -0.0743 | No |
| 59 | Tap1 | 9806 | -0.005 | -0.0814 | No |
| 60 | Ykt6 | 10049 | -0.013 | -0.0936 | No |
| 61 | Ddx21 | 10184 | -0.018 | -0.1000 | No |
| 62 | Rpn1 | 10449 | -0.027 | -0.1128 | No |
| 63 | Rhob | 10460 | -0.027 | -0.1124 | No |
| 64 | Fen1 | 10616 | -0.032 | -0.1194 | No |
| 65 | Klhdc3 | 10673 | -0.034 | -0.1211 | No |
| 66 | Gls | 10743 | -0.036 | -0.1235 | No |
| 67 | Casp3 | 11057 | -0.047 | -0.1382 | No |
| 68 | Mark2 | 11111 | -0.049 | -0.1392 | No |
| 69 | Dnaja1 | 11303 | -0.055 | -0.1473 | No |
| 70 | Hnrnpu | 11526 | -0.064 | -0.1567 | No |
| 71 | Prpf3 | 11559 | -0.065 | -0.1561 | No |
| 72 | Creg1 | 11817 | -0.074 | -0.1669 | No |
| 73 | Eno2 | 12230 | -0.090 | -0.1853 | No |
| 74 | Dgat1 | 12337 | -0.094 | -0.1876 | No |
| 75 | Aldoa | 12356 | -0.094 | -0.1853 | No |
| 76 | Amd1 | 12419 | -0.096 | -0.1852 | No |
| 77 | Kcnh2 | 12452 | -0.098 | -0.1835 | No |
| 78 | Ephx1 | 12482 | -0.099 | -0.1816 | No |
| 79 | Car2 | 12485 | -0.099 | -0.1783 | No |
| 80 | Selenow | 12585 | -0.104 | -0.1799 | No |
| 81 | Plcl1 | 12739 | -0.110 | -0.1841 | No |
| 82 | Atf3 | 12989 | -0.119 | -0.1930 | No |
| 83 | Mmp14 | 13144 | -0.123 | -0.1968 | No |
| 84 | Stip1 | 13175 | -0.124 | -0.1940 | No |
| 85 | Rrad | 13241 | -0.127 | -0.1930 | No |
| 86 | Ptprd | 13628 | -0.142 | -0.2083 | No |
| 87 | Parp2 | 13776 | -0.149 | -0.2108 | No |
| 88 | Tyro3 | 13927 | -0.156 | -0.2132 | No |
| 89 | Maoa | 13972 | -0.158 | -0.2101 | No |
| 90 | Dlg4 | 14105 | -0.163 | -0.2113 | No |
| 91 | Acaa1a | 14127 | -0.164 | -0.2068 | No |
| 92 | Lyn | 14419 | -0.177 | -0.2159 | Yes |
| 93 | Fosb | 14426 | -0.177 | -0.2101 | Yes |
| 94 | H2ax | 14531 | -0.182 | -0.2092 | Yes |
| 95 | Prkcd | 14606 | -0.185 | -0.2067 | Yes |
| 96 | Ppt1 | 14693 | -0.189 | -0.2047 | Yes |
| 97 | Olfm1 | 14758 | -0.192 | -0.2014 | Yes |
| 98 | Gch1 | 14855 | -0.196 | -0.1996 | Yes |
| 99 | Htr7 | 15026 | -0.206 | -0.2014 | Yes |
| 100 | Cdo1 | 15169 | -0.213 | -0.2015 | Yes |
| 101 | Ggh | 15193 | -0.214 | -0.1953 | Yes |
| 102 | Mrpl23 | 15207 | -0.215 | -0.1886 | Yes |
| 103 | Hspa13 | 15223 | -0.215 | -0.1819 | Yes |
| 104 | Btg3 | 15259 | -0.217 | -0.1763 | Yes |
| 105 | Wiz | 15387 | -0.225 | -0.1751 | Yes |
| 106 | Btg2 | 15551 | -0.232 | -0.1756 | Yes |
| 107 | Col2a1 | 15699 | -0.241 | -0.1750 | Yes |
| 108 | Psmc3 | 15740 | -0.242 | -0.1687 | Yes |
| 109 | Bmp2 | 15845 | -0.247 | -0.1656 | Yes |
| 110 | Cyb5r1 | 16043 | -0.260 | -0.1669 | Yes |
| 111 | Junb | 16128 | -0.266 | -0.1621 | Yes |
| 112 | Cyb5b | 16242 | -0.273 | -0.1586 | Yes |
| 113 | Nfkbia | 16280 | -0.275 | -0.1510 | Yes |
| 114 | Pole3 | 16398 | -0.283 | -0.1473 | Yes |
| 115 | Asns | 16667 | -0.302 | -0.1509 | Yes |
| 116 | Cdc34 | 16677 | -0.302 | -0.1410 | Yes |
| 117 | Tfrc | 16751 | -0.306 | -0.1342 | Yes |
| 118 | Cebpg | 16819 | -0.310 | -0.1270 | Yes |
| 119 | Il6st | 17027 | -0.328 | -0.1265 | Yes |
| 120 | Atp6v1f | 17334 | -0.353 | -0.1302 | Yes |
| 121 | Mgat1 | 17381 | -0.358 | -0.1203 | Yes |
| 122 | Urod | 17551 | -0.371 | -0.1163 | Yes |
| 123 | Ret | 17597 | -0.376 | -0.1057 | Yes |
| 124 | Eif2s3x | 17943 | -0.410 | -0.1095 | Yes |
| 125 | Stk25 | 17969 | -0.412 | -0.0966 | Yes |
| 126 | Irf1 | 18157 | -0.434 | -0.0914 | Yes |
| 127 | Ap2s1 | 18284 | -0.451 | -0.0824 | Yes |
| 128 | Cnp | 18389 | -0.466 | -0.0718 | Yes |
| 129 | Ntrk3 | 18392 | -0.467 | -0.0558 | Yes |
| 130 | Ccnd3 | 18595 | -0.505 | -0.0489 | Yes |
| 131 | Icam1 | 18721 | -0.534 | -0.0370 | Yes |
| 132 | E2f5 | 18900 | -0.580 | -0.0262 | Yes |
| 133 | Sqstm1 | 18967 | -0.605 | -0.0088 | Yes |
| 134 | Il6 | 19208 | -0.767 | 0.0052 | Yes |