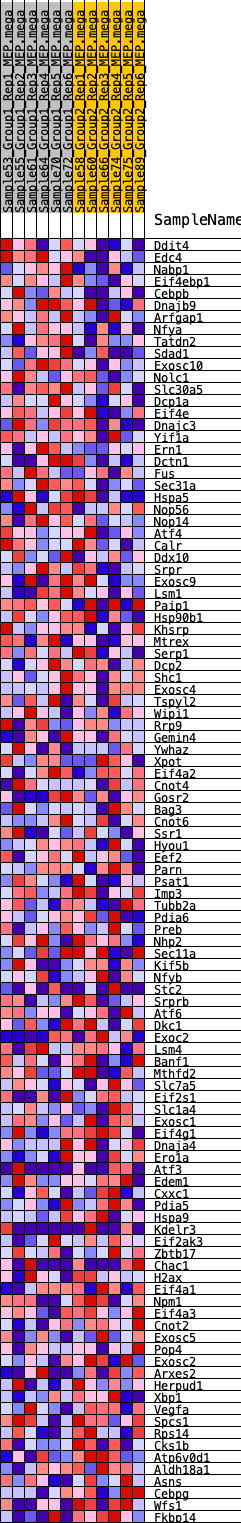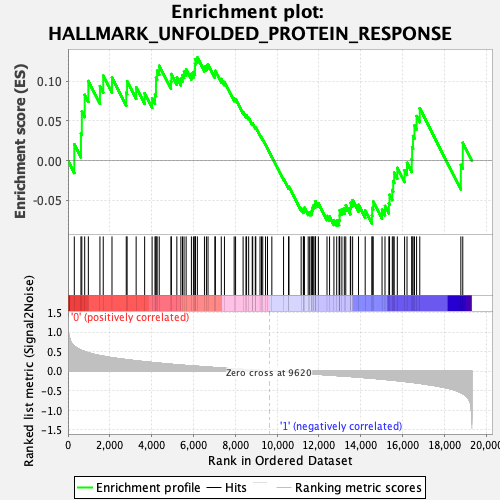
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_UNFOLDED_PROTEIN_RESPONSE |
| Enrichment Score (ES) | 0.12984927 |
| Normalized Enrichment Score (NES) | 0.56584036 |
| Nominal p-value | 0.9826255 |
| FDR q-value | 0.97297436 |
| FWER p-Value | 1.0 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Ddit4 | 301 | 0.639 | 0.0205 | Yes |
| 2 | Edc4 | 619 | 0.533 | 0.0342 | Yes |
| 3 | Nabp1 | 661 | 0.525 | 0.0618 | Yes |
| 4 | Eif4ebp1 | 798 | 0.498 | 0.0829 | Yes |
| 5 | Cebpb | 973 | 0.467 | 0.1002 | Yes |
| 6 | Dnajb9 | 1527 | 0.395 | 0.0938 | Yes |
| 7 | Arfgap1 | 1685 | 0.379 | 0.1071 | Yes |
| 8 | Nfya | 2103 | 0.340 | 0.1046 | Yes |
| 9 | Tatdn2 | 2785 | 0.292 | 0.0857 | Yes |
| 10 | Sdad1 | 2826 | 0.290 | 0.1000 | Yes |
| 11 | Exosc10 | 3258 | 0.262 | 0.0924 | Yes |
| 12 | Nolc1 | 3664 | 0.237 | 0.0847 | Yes |
| 13 | Slc30a5 | 4021 | 0.219 | 0.0786 | Yes |
| 14 | Dcp1a | 4160 | 0.211 | 0.0833 | Yes |
| 15 | Eif4e | 4206 | 0.208 | 0.0927 | Yes |
| 16 | Dnajc3 | 4208 | 0.207 | 0.1044 | Yes |
| 17 | Yif1a | 4262 | 0.205 | 0.1133 | Yes |
| 18 | Ern1 | 4362 | 0.201 | 0.1195 | Yes |
| 19 | Dctn1 | 4923 | 0.174 | 0.1002 | Yes |
| 20 | Fus | 4942 | 0.173 | 0.1091 | Yes |
| 21 | Sec31a | 5204 | 0.161 | 0.1046 | Yes |
| 22 | Hspa5 | 5397 | 0.152 | 0.1032 | Yes |
| 23 | Nop56 | 5474 | 0.149 | 0.1076 | Yes |
| 24 | Nop14 | 5548 | 0.146 | 0.1121 | Yes |
| 25 | Atf4 | 5648 | 0.142 | 0.1150 | Yes |
| 26 | Calr | 5900 | 0.131 | 0.1093 | Yes |
| 27 | Ddx10 | 5997 | 0.127 | 0.1115 | Yes |
| 28 | Srpr | 6070 | 0.125 | 0.1148 | Yes |
| 29 | Exosc9 | 6073 | 0.125 | 0.1218 | Yes |
| 30 | Lsm1 | 6094 | 0.124 | 0.1278 | Yes |
| 31 | Paip1 | 6185 | 0.120 | 0.1298 | Yes |
| 32 | Hsp90b1 | 6524 | 0.108 | 0.1184 | No |
| 33 | Khsrp | 6618 | 0.104 | 0.1194 | No |
| 34 | Mtrex | 6695 | 0.101 | 0.1211 | No |
| 35 | Serp1 | 7023 | 0.089 | 0.1092 | No |
| 36 | Dcp2 | 7046 | 0.088 | 0.1130 | No |
| 37 | Shc1 | 7328 | 0.078 | 0.1028 | No |
| 38 | Exosc4 | 7480 | 0.072 | 0.0990 | No |
| 39 | Tspyl2 | 7945 | 0.056 | 0.0780 | No |
| 40 | Wipi1 | 8012 | 0.053 | 0.0776 | No |
| 41 | Rrp9 | 8372 | 0.042 | 0.0612 | No |
| 42 | Gemin4 | 8508 | 0.038 | 0.0563 | No |
| 43 | Ywhaz | 8538 | 0.037 | 0.0569 | No |
| 44 | Xpot | 8638 | 0.034 | 0.0537 | No |
| 45 | Eif4a2 | 8809 | 0.027 | 0.0464 | No |
| 46 | Cnot4 | 8831 | 0.027 | 0.0468 | No |
| 47 | Gosr2 | 8955 | 0.022 | 0.0416 | No |
| 48 | Bag3 | 8979 | 0.021 | 0.0416 | No |
| 49 | Cnot6 | 9181 | 0.015 | 0.0320 | No |
| 50 | Ssr1 | 9252 | 0.012 | 0.0290 | No |
| 51 | Hyou1 | 9311 | 0.010 | 0.0266 | No |
| 52 | Eef2 | 9445 | 0.006 | 0.0200 | No |
| 53 | Parn | 9538 | 0.003 | 0.0154 | No |
| 54 | Psat1 | 9746 | -0.003 | 0.0048 | No |
| 55 | Imp3 | 10306 | -0.022 | -0.0231 | No |
| 56 | Tubb2a | 10545 | -0.030 | -0.0338 | No |
| 57 | Pdia6 | 10564 | -0.031 | -0.0330 | No |
| 58 | Preb | 11147 | -0.050 | -0.0604 | No |
| 59 | Nhp2 | 11253 | -0.054 | -0.0629 | No |
| 60 | Sec11a | 11264 | -0.054 | -0.0603 | No |
| 61 | Kif5b | 11304 | -0.055 | -0.0592 | No |
| 62 | Nfyb | 11484 | -0.062 | -0.0650 | No |
| 63 | Stc2 | 11554 | -0.065 | -0.0649 | No |
| 64 | Srprb | 11628 | -0.068 | -0.0649 | No |
| 65 | Atf6 | 11663 | -0.069 | -0.0628 | No |
| 66 | Dkc1 | 11685 | -0.069 | -0.0600 | No |
| 67 | Exoc2 | 11717 | -0.070 | -0.0576 | No |
| 68 | Lsm4 | 11760 | -0.072 | -0.0557 | No |
| 69 | Banf1 | 11827 | -0.074 | -0.0550 | No |
| 70 | Mthfd2 | 11836 | -0.075 | -0.0512 | No |
| 71 | Slc7a5 | 11970 | -0.081 | -0.0535 | No |
| 72 | Eif2s1 | 12383 | -0.095 | -0.0696 | No |
| 73 | Slc1a4 | 12506 | -0.100 | -0.0703 | No |
| 74 | Exosc1 | 12714 | -0.109 | -0.0749 | No |
| 75 | Eif4g1 | 12847 | -0.113 | -0.0754 | No |
| 76 | Dnaja4 | 12962 | -0.118 | -0.0747 | No |
| 77 | Ero1a | 12987 | -0.119 | -0.0692 | No |
| 78 | Atf3 | 12989 | -0.119 | -0.0625 | No |
| 79 | Edem1 | 13096 | -0.122 | -0.0612 | No |
| 80 | Cxxc1 | 13211 | -0.126 | -0.0600 | No |
| 81 | Pdia5 | 13281 | -0.129 | -0.0563 | No |
| 82 | Hspa9 | 13499 | -0.137 | -0.0599 | No |
| 83 | Kdelr3 | 13516 | -0.137 | -0.0529 | No |
| 84 | Eif2ak3 | 13609 | -0.142 | -0.0497 | No |
| 85 | Zbtb17 | 13890 | -0.154 | -0.0555 | No |
| 86 | Chac1 | 14209 | -0.168 | -0.0626 | No |
| 87 | H2ax | 14531 | -0.182 | -0.0690 | No |
| 88 | Eif4a1 | 14545 | -0.183 | -0.0593 | No |
| 89 | Npm1 | 14593 | -0.184 | -0.0514 | No |
| 90 | Eif4a3 | 15015 | -0.205 | -0.0617 | No |
| 91 | Cnot2 | 15156 | -0.212 | -0.0570 | No |
| 92 | Exosc5 | 15341 | -0.222 | -0.0540 | No |
| 93 | Pop4 | 15373 | -0.224 | -0.0430 | No |
| 94 | Exosc2 | 15506 | -0.230 | -0.0368 | No |
| 95 | Arxes2 | 15549 | -0.232 | -0.0259 | No |
| 96 | Herpud1 | 15596 | -0.235 | -0.0150 | No |
| 97 | Xbp1 | 15745 | -0.243 | -0.0090 | No |
| 98 | Vegfa | 16093 | -0.264 | -0.0121 | No |
| 99 | Spcs1 | 16209 | -0.272 | -0.0027 | No |
| 100 | Rps14 | 16435 | -0.285 | 0.0017 | No |
| 101 | Cks1b | 16453 | -0.287 | 0.0171 | No |
| 102 | Atp6v0d1 | 16499 | -0.289 | 0.0311 | No |
| 103 | Aldh18a1 | 16566 | -0.294 | 0.0443 | No |
| 104 | Asns | 16667 | -0.302 | 0.0562 | No |
| 105 | Cebpg | 16819 | -0.310 | 0.0659 | No |
| 106 | Wfs1 | 18783 | -0.549 | -0.0053 | No |
| 107 | Fkbp14 | 18875 | -0.575 | 0.0225 | No |