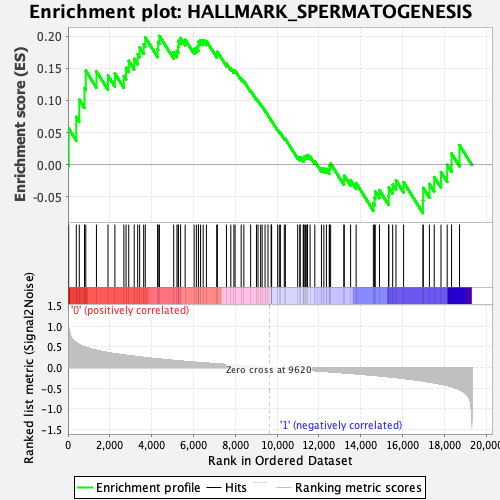
Profile of the Running ES Score & Positions of GeneSet Members on the Rank Ordered List
| Dataset | MEP.MEP.mega_Pheno.cls #Group1_versus_Group2.MEP.mega_Pheno.cls #Group1_versus_Group2_repos |
| Phenotype | MEP.mega_Pheno.cls#Group1_versus_Group2_repos |
| Upregulated in class | 0 |
| GeneSet | HALLMARK_SPERMATOGENESIS |
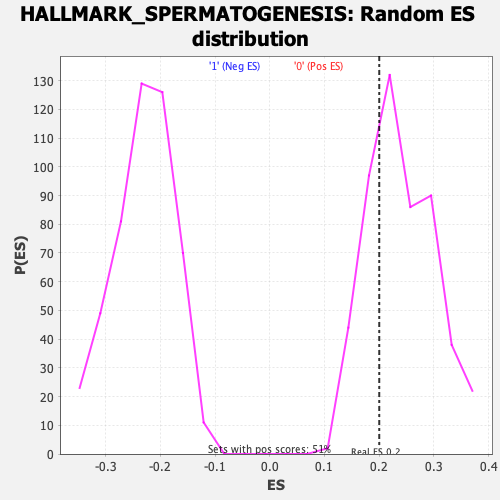
| Enrichment Score (ES) | 0.20022687 |
| Normalized Enrichment Score (NES) | 0.83601975 |
| Nominal p-value | 0.72015655 |
| FDR q-value | 1.0 |
| FWER p-Value | 0.997 |

| SYMBOL | RANK IN GENE LIST | RANK METRIC SCORE | RUNNING ES | CORE ENRICHMENT | |
|---|---|---|---|---|---|
| 1 | Septin4 | 33 | 0.945 | 0.0565 | Yes |
| 2 | Gfi1 | 397 | 0.598 | 0.0745 | Yes |
| 3 | Cftr | 539 | 0.555 | 0.1013 | Yes |
| 4 | Il12rb2 | 789 | 0.501 | 0.1192 | Yes |
| 5 | Dmc1 | 850 | 0.489 | 0.1462 | Yes |
| 6 | Gstm5 | 1360 | 0.412 | 0.1451 | Yes |
| 7 | Mast2 | 1912 | 0.358 | 0.1385 | Yes |
| 8 | Clvs1 | 2242 | 0.331 | 0.1418 | Yes |
| 9 | Agfg1 | 2672 | 0.300 | 0.1379 | Yes |
| 10 | Ybx2 | 2776 | 0.293 | 0.1506 | Yes |
| 11 | Rfc4 | 2904 | 0.284 | 0.1615 | Yes |
| 12 | Topbp1 | 3164 | 0.269 | 0.1646 | Yes |
| 13 | Mtor | 3332 | 0.257 | 0.1717 | Yes |
| 14 | Grm8 | 3425 | 0.252 | 0.1825 | Yes |
| 15 | Tnni3 | 3618 | 0.240 | 0.1873 | Yes |
| 16 | Prkar2a | 3694 | 0.235 | 0.1979 | Yes |
| 17 | Ide | 4284 | 0.205 | 0.1798 | Yes |
| 18 | Oaz3 | 4310 | 0.204 | 0.1911 | Yes |
| 19 | Gmcl1 | 4372 | 0.200 | 0.2002 | Yes |
| 20 | Slc2a5 | 5054 | 0.168 | 0.1751 | No |
| 21 | Crisp2 | 5209 | 0.160 | 0.1770 | No |
| 22 | Hspa2 | 5270 | 0.158 | 0.1836 | No |
| 23 | Pcsk4 | 5280 | 0.157 | 0.1928 | No |
| 24 | Adam2 | 5383 | 0.152 | 0.1969 | No |
| 25 | Phf7 | 5601 | 0.144 | 0.1945 | No |
| 26 | Ift88 | 6029 | 0.126 | 0.1800 | No |
| 27 | Arl4a | 6137 | 0.122 | 0.1820 | No |
| 28 | Stam2 | 6236 | 0.119 | 0.1842 | No |
| 29 | Mllt10 | 6238 | 0.119 | 0.1915 | No |
| 30 | Zc3h14 | 6338 | 0.115 | 0.1935 | No |
| 31 | Ncaph | 6463 | 0.110 | 0.1938 | No |
| 32 | Nf2 | 6617 | 0.104 | 0.1923 | No |
| 33 | Psmg1 | 7110 | 0.086 | 0.1719 | No |
| 34 | Pgs1 | 7142 | 0.085 | 0.1756 | No |
| 35 | Coil | 7576 | 0.069 | 0.1573 | No |
| 36 | Art3 | 7785 | 0.062 | 0.1502 | No |
| 37 | Hspa4l | 7934 | 0.056 | 0.1460 | No |
| 38 | Slc12a2 | 7988 | 0.054 | 0.1466 | No |
| 39 | Ezh2 | 8280 | 0.044 | 0.1342 | No |
| 40 | Spata6 | 8408 | 0.041 | 0.1301 | No |
| 41 | Mlf1 | 8732 | 0.031 | 0.1152 | No |
| 42 | Bub1 | 9011 | 0.020 | 0.1019 | No |
| 43 | Nefh | 9085 | 0.018 | 0.0993 | No |
| 44 | Camk4 | 9211 | 0.014 | 0.0936 | No |
| 45 | Nphp1 | 9282 | 0.011 | 0.0907 | No |
| 46 | She | 9422 | 0.007 | 0.0838 | No |
| 47 | Gad1 | 9562 | 0.002 | 0.0767 | No |
| 48 | Strbp | 9712 | -0.002 | 0.0691 | No |
| 49 | Acrbp | 9729 | -0.002 | 0.0684 | No |
| 50 | Ddx25 | 9736 | -0.003 | 0.0683 | No |
| 51 | Zpbp | 10024 | -0.012 | 0.0541 | No |
| 52 | Dbf4 | 10103 | -0.015 | 0.0510 | No |
| 53 | Ddx4 | 10155 | -0.017 | 0.0494 | No |
| 54 | Braf | 10342 | -0.023 | 0.0411 | No |
| 55 | Pacrg | 10393 | -0.025 | 0.0400 | No |
| 56 | Jam3 | 10981 | -0.044 | 0.0122 | No |
| 57 | Vdac3 | 11082 | -0.048 | 0.0099 | No |
| 58 | Pomc | 11101 | -0.049 | 0.0120 | No |
| 59 | Pias2 | 11247 | -0.054 | 0.0078 | No |
| 60 | Cdkn3 | 11277 | -0.054 | 0.0096 | No |
| 61 | Tsn | 11286 | -0.055 | 0.0126 | No |
| 62 | Cct6b | 11359 | -0.057 | 0.0124 | No |
| 63 | Pebp1 | 11416 | -0.060 | 0.0131 | No |
| 64 | Chfr | 11451 | -0.061 | 0.0152 | No |
| 65 | Zc2hc1c | 11575 | -0.066 | 0.0128 | No |
| 66 | Scg5 | 11800 | -0.073 | 0.0056 | No |
| 67 | Sirt1 | 12118 | -0.086 | -0.0055 | No |
| 68 | Dmrt1 | 12233 | -0.090 | -0.0059 | No |
| 69 | Cnih2 | 12355 | -0.094 | -0.0064 | No |
| 70 | Taldo1 | 12491 | -0.099 | -0.0073 | No |
| 71 | Lpin1 | 12503 | -0.100 | -0.0017 | No |
| 72 | Tle4 | 12557 | -0.102 | 0.0018 | No |
| 73 | Map7 | 13187 | -0.125 | -0.0232 | No |
| 74 | Ccnb2 | 13209 | -0.126 | -0.0166 | No |
| 75 | Csnk2a2 | 13512 | -0.137 | -0.0238 | No |
| 76 | Parp2 | 13776 | -0.149 | -0.0283 | No |
| 77 | Tekt2 | 14600 | -0.184 | -0.0598 | No |
| 78 | Ip6k1 | 14660 | -0.187 | -0.0513 | No |
| 79 | Nos1 | 14689 | -0.188 | -0.0412 | No |
| 80 | Phkg2 | 14893 | -0.198 | -0.0395 | No |
| 81 | Tulp2 | 15329 | -0.221 | -0.0486 | No |
| 82 | Gapdhs | 15338 | -0.221 | -0.0353 | No |
| 83 | Sycp1 | 15523 | -0.231 | -0.0307 | No |
| 84 | Cdk1 | 15681 | -0.240 | -0.0241 | No |
| 85 | Adad1 | 16046 | -0.260 | -0.0270 | No |
| 86 | Dcc | 16971 | -0.323 | -0.0552 | No |
| 87 | Kif2c | 16985 | -0.325 | -0.0358 | No |
| 88 | Ttk | 17280 | -0.348 | -0.0297 | No |
| 89 | Clpb | 17510 | -0.367 | -0.0190 | No |
| 90 | Rad17 | 17833 | -0.399 | -0.0111 | No |
| 91 | Aurka | 18130 | -0.431 | -0.0000 | No |
| 92 | Tcp11 | 18339 | -0.459 | 0.0174 | No |
| 93 | Nek2 | 18720 | -0.534 | 0.0305 | No |